Week 4: Data manipulation and statistical analysis
ACTL1101 Introduction to Actuarial Studies
Learning outcomes
By the end of this topic, you should be able to
- manipulate different data structures using appropriate functions
- rectify incorrectly imported data
- summarise and organise data in a variety of ways, including with package
dplyr - compute useful statistics from imported data
Data frames refresher
Data frames - Exercise
Let’s start with an exercise using data frames to remind ourselves how they work.
Consider some data stored in a data frame:
- What is the sum of everyone’s heights?
- What are the full details of James? (Try to return the entire row that corresponds to James)
- What is the average income of the males?
- Who is the shortest person? Hint: use
which.min
Aside on this week
- You may have noticed that even for simple examples as those of the previous slide, R syntax can get clunky. You are not alone.
- But there is good news. In the last section of this week’s slides, we learn about a new R syntax that makes manipulating data easier.
- Indeed, we will use the external package
dplyr, which is part of the tidyverse. This is a collection of R packages for data science that aim to make R more powerful and easy to use. Stay tuned.
The attach() function
The function
attach(some.data.frame)creates a workspace where each variable of the data frame can be accessed directly (without using the dollar sign$). Therefore it can reduce the length of your code. However, these “attached” columns now exist “independently” of the data frame. In other words, if you make a change in the actual data frame, it will not affect the attached columns.We recommend to be very careful with the
attach()function (especially for large projects).
Statistical analysis
Importing data - Pages 339-343
Here we use the sample_data.csv data to illustrate a few handy statistical functions. This data is already in Ed (in this week’s “Lab Exercise Space”). You can download it if you want to use it on your computer.
- import the data to R (if doing this on your computer, first save the data in the folder you set as the working directory)
- use the
attach()function to gain direct access to the variables
dob gender situation tea coffee height weight age meat fish
1 1967-01-28 Female single 0 0 151 58 58 4-6/week. 2-3/week.
2 1965-05-05 Female single 1 1 162 60 60 1/day. 1/week.
3 1950-11-21 Female single 0 4 162 75 75 2-3/week. <1/week.
4 1980-03-16 Female single 0 0 154 45 45 never 4-6/week.
5 1975-07-20 Female single 2 1 154 50 50 1/day. 2-3/week.
6 1959-07-23 Female single 2 0 159 66 66 4-6/week. 1/week.
raw_fruit cooked_fruit_veg chocol fat
1 <1/week. 4-6/week. 1/day. Isio4
2 1/day. 1/day. <1/week. sunflower
3 1/day. 1/week. 1/day. sunflower
4 4-6/week. never 2-3/week. margarine
5 1/day. 1/day. 2-3/week. margarine
6 1/day. 1/day. <1/week. peanutStatistical Analysis - Variables - Pages 339-343
\(14\) variables (columns) of \(226\) individuals (rows)
qualitative:
gender,fatandsituationordinal:
meat,fish,raw_fruit,cooked_fruit_vegandchocoldiscrete quantitative:
tea,coffeecontinuous quantitative:
height,weightandageother:
dob(dates do not really fit into any category cleanly, and their qualification depends on the use case)
Notes
The
sample_data.csvis created based from thenutrition_elderly.csvspreadsheet downloaded from http://www.biostatisticien.eu/springeR/nutrition_elderly.xls. See Page 339.Variables of
nutrition_elderly.csvhave been structured following the steps from Pages 340-343 and a new data frame was created and saved assample_data.csv
Numerical statistics - Pages 347-352
Some useful statistics for a preliminary analysis (of variable weight in this example):
[1] 66.4823[1] 66[1] 12.03337[1] 58[1] 17.75[1] 9.919884 10% 90%
51.5 82.0 Note: Remember, variable weight can be accessed directly only because we use the command attach(sample_data) previously!
Numerical statistics
While knowing all of these functions is helpful for individual variables, it is often useful to get an overall picture of the dataset, and for this we have the summary() function.
dob gender situation tea
Length:226 Length:226 Length:226 Min. : 0.0000
Class :character Class :character Class :character 1st Qu.: 0.0000
Mode :character Mode :character Mode :character Median : 0.0000
Mean : 0.7124
3rd Qu.: 1.0000
Max. :10.0000
coffee height weight age
Min. :0.000 Min. :140 Min. :38.00 Min. :38.00
1st Qu.:1.000 1st Qu.:157 1st Qu.:57.25 1st Qu.:57.25
Median :2.000 Median :163 Median :66.00 Median :66.00
Mean :1.619 Mean :164 Mean :66.48 Mean :66.48
3rd Qu.:2.000 3rd Qu.:170 3rd Qu.:75.00 3rd Qu.:75.00
Max. :5.000 Max. :188 Max. :96.00 Max. :96.00
meat fish raw_fruit cooked_fruit_veg
Length:226 Length:226 Length:226 Length:226
Class :character Class :character Class :character Class :character
Mode :character Mode :character Mode :character Mode :character
chocol fat
Length:226 Length:226
Class :character Class :character
Mode :character Mode :character
Do you notice something strange? There are some issues here:
- Variables like
genderare not being treated as qualitative, but as arbitrary characters. - Likewise, variables like
fishandmeatare being treated as arbitrary characters instead of ordinal. - Likewise,
dobis being treated as arbitrary characters, instead of a date.
We also could have found these issues by using str(sample_data).
Correcting the qualitative/ordinal variables
When importing external data, it is common to have issues like those on the previous slide, so it is worthwhile to look at how we can fix those problems.
- We can use
factorfor this. - We can use
factorwithordered=Tandlevels(we saw this in week 2). Rhas a specific data structure for dates. We can convert acharacterto adatewith functionas.Date.
Starting with the first two
The other variables have been corrected as well but omitted here for brevity. You should try to construct them yourself.
Correcting the dates
- To convert dates we use the
as.Datefunction. Of course, dates can be stored in many formats (e.g."15-06-2025","15/06/25","2025-Jun-15"etc), and we must tellRwhich format we have, using theformatargument.
[1] "1967-01-28" "1965-05-05" "1950-11-21"The format specified above says we are looking for a 4 digit year with the month and day represented by numbers. You can see a full list of format specifiers (with some more advanced use cases) here.
While it may not be immediately obvious why using the proper
datestructure is useful, to perform visualisation or complex manipulations, having “proper” date objects is required. To give you a small use case, note that you can “add” to a date, but not if it’s stored as a character.
[1] "2025-07-18"[1] "2025-07-25"Error in "2025-06-21" + 7: non-numeric argument to binary operator- One last note:
Rstores dates in the formatYYYY-mm-dd. You can display a date in another format using functionformat. However, when doing this the date reverts back to acharacter.
Checking again our imported data
Once this is all done we can view the summary, which is now much more informative.
dob gender situation tea
Min. :1929-01-09 Female:141 couple:119 Min. : 0.0000
1st Qu.:1950-06-24 Male : 85 family: 9 1st Qu.: 0.0000
Median :1959-08-30 single: 98 Median : 0.0000
Mean :1959-01-12 Mean : 0.7124
3rd Qu.:1968-03-31 3rd Qu.: 1.0000
Max. :1987-05-14 Max. :10.0000
coffee height weight age meat
Min. :0.000 Min. :140 Min. :38.00 Min. :38.00 never : 1
1st Qu.:1.000 1st Qu.:157 1st Qu.:57.25 1st Qu.:57.25 <1/week. : 3
Median :2.000 Median :163 Median :66.00 Median :66.00 1/week. :11
Mean :1.619 Mean :164 Mean :66.48 Mean :66.48 2-3/week.:83
3rd Qu.:2.000 3rd Qu.:170 3rd Qu.:75.00 3rd Qu.:75.00 4-6/week.:67
Max. :5.000 Max. :188 Max. :96.00 Max. :96.00 1/day. :61
fish raw_fruit cooked_fruit_veg chocol
never : 4 never : 2 never : 2 never :50
<1/week. : 21 <1/week. : 8 <1/week. : 3 <1/week. :62
1/week. : 61 1/week. : 8 1/week. : 7 1/week. :16
2-3/week.:118 2-3/week.: 14 2-3/week.: 30 2-3/week.:22
4-6/week.: 15 4-6/week.: 22 4-6/week.: 36 4-6/week.:11
1/day. : 7 1/day. :172 1/day. :148 1/day. :65
fat
sunflower:68
peanut :48
olive :40
margarine:27
Isio4 :23
butter :15
(Other) : 5 - A good data analyst always checks the format and quality of the data they imported.
- While the previous slides have shown a few simple cases, there are many more potential issues you can (will!) encounter when importing data.
- There is no escaping it: albeit “boring”, cleaning up these issues will often constitute a large part of your work as a data analyst.
Frequency table - discrete variables - Pages 343-344
For qualitative variable situation:
Frequency table - joint observations - Pages 344-345
For the paired observations of gender and situation:
situation
gender couple family single
Female 56 7 78
Male 63 2 20Margins computed over dimensions
in the following order:
1: gender
2: situation situation
gender couple family single sum
Female 56 7 78 141
Male 63 2 20 85
sum 119 9 98 226Correlation: an association measure - Pages 354-355
The cor() function computes the (sample) correlation between two variables, which is a measure of linear association. As an example, we compute the correlation between two variables from the in-built dataset mtcars:
wt: weight of the car (in 1000’s of pounds)mgp: miles per gallon
The correlation always ranges from \(-1\) to \(1\). Here a correlation of \(-0.868\) indicates a strong negative association between wt and mgp: the heavier the car, the less distance it can go with one gallon of fuel (which makes sense!).
#Plot one variable vs the other
plot(mtcars$wt, mtcars$mpg, main = "Miles per gallon vs Weight of a car",
xlab="weight",ylab ="miles/gallon")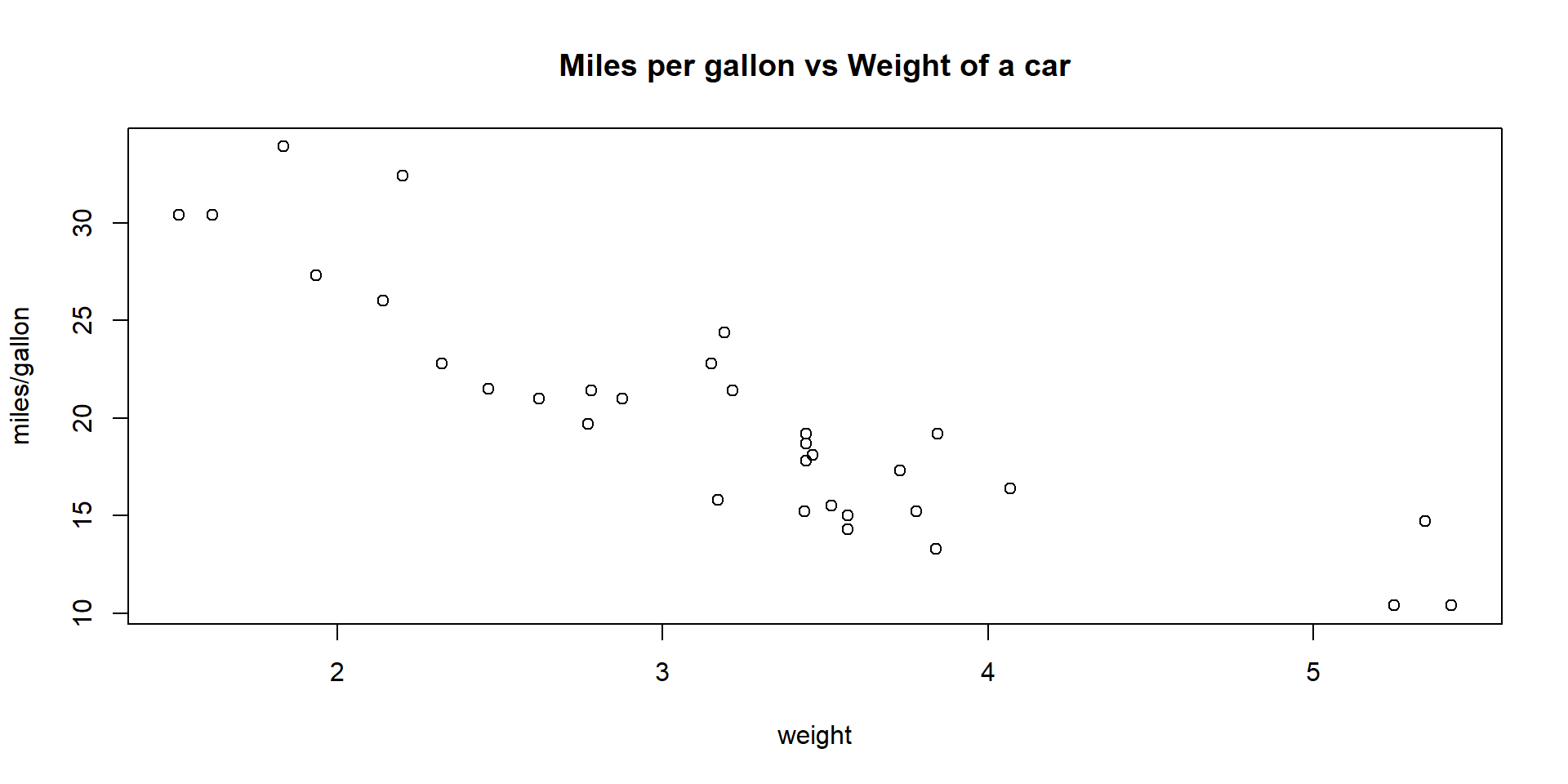
Note: We will see more about correlation in Module 4: Risk & Insurance (Theory component of the course).
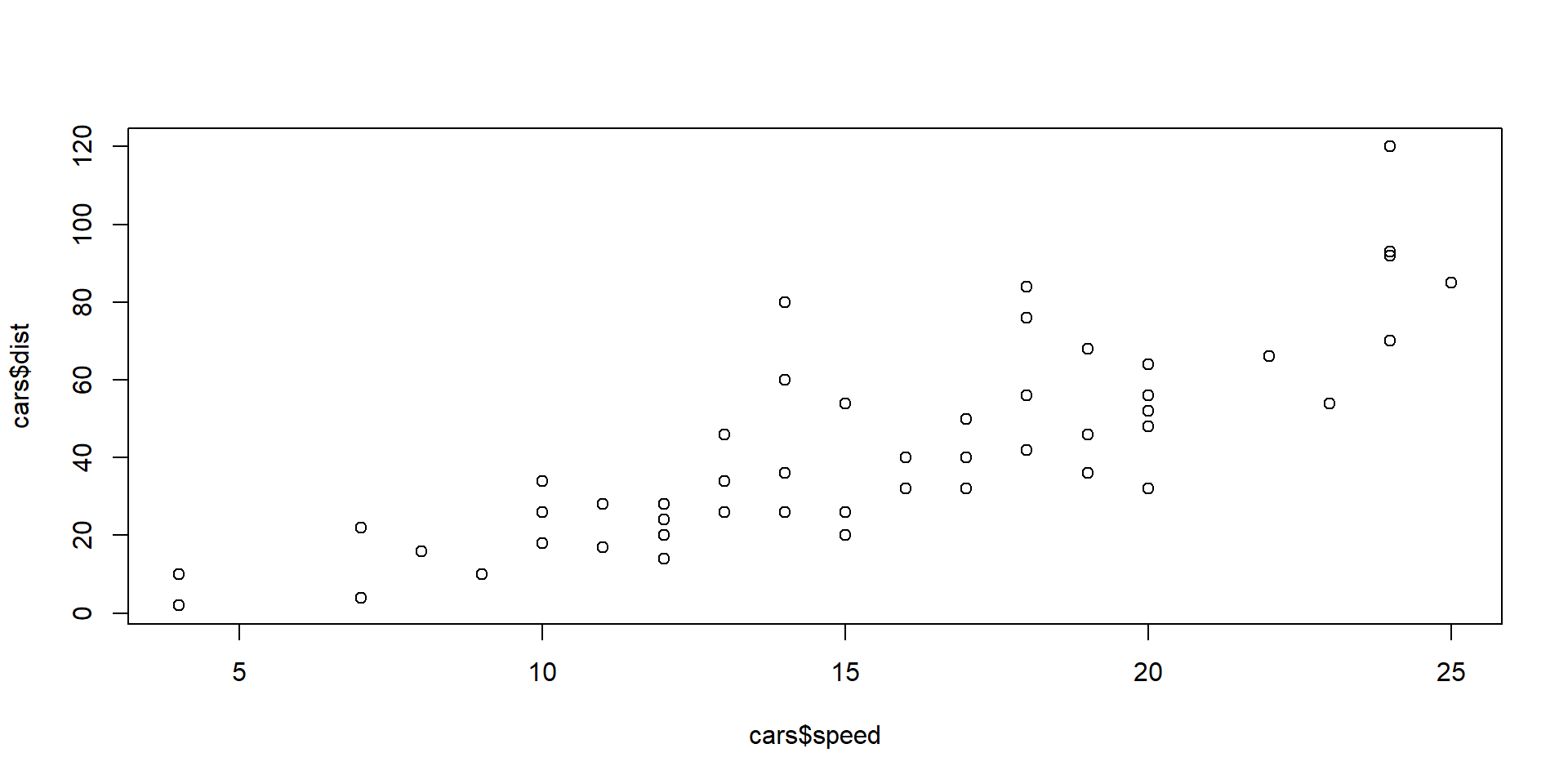
Exercise
Use the in-built R dataset cars. Calculate the correlation between the speed and dist (for stopping distance) from the 50 observations.
What can you learn? Is there any further investigation you want to do?
Solution
Comments:
The correlation is \(0.807\), which indicates a high level of positive linear association between the two variables, i.e. if one variable is “high”, the other variable also tends to be “high”.
We can create the scatter plot of one variable versus the other and examine the relationship ‘visually’. More on plots next Week!
Here it turns out that an even better description of the relationship between these two variables is a linear relationship between the log-scaled values… also more on that later!
dplyr
dplyr - Introduction
dplyr is a commonly used package (which we need to load into R as it is not part of ‘base R’). It makes manipulating dataframes very easy and intuitive and is great for data analysis. To install any package, just type
- To use a specific function from a package, the syntax is
dplyr::function_name(). - This is okay if you use a function once or twice, but in general it is better to load the entire package into our workspace:
You can then use all functions in the package, for as long as your R session lasts. There are 6 main functions in
dplyr:filter- selects rowsselect- selects columnsarrange- sorts rows by some ordermutate- creates new columnsgroup_by- groups certain rows togethersummarise- provides summaries of grouped rows
dplyr - Filter and Select
Here some examples using the in-built data frame mtcars
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 mpg cyl disp hp drat wt qsec vs am gear carb
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2 mpg cyl wt
Mazda RX4 21.0 6 2.620
Mazda RX4 Wag 21.0 6 2.875
Datsun 710 22.8 4 2.320
Hornet 4 Drive 21.4 6 3.215
Hornet Sportabout 18.7 8 3.440
Valiant 18.1 6 3.460
Duster 360 14.3 8 3.570
Merc 240D 24.4 4 3.190
Merc 230 22.8 4 3.150
Merc 280 19.2 6 3.440
Merc 280C 17.8 6 3.440
Merc 450SE 16.4 8 4.070
Merc 450SL 17.3 8 3.730
Merc 450SLC 15.2 8 3.780
Cadillac Fleetwood 10.4 8 5.250
Lincoln Continental 10.4 8 5.424
Chrysler Imperial 14.7 8 5.345
Fiat 128 32.4 4 2.200
Honda Civic 30.4 4 1.615
Toyota Corolla 33.9 4 1.835
Toyota Corona 21.5 4 2.465
Dodge Challenger 15.5 8 3.520
AMC Javelin 15.2 8 3.435
Camaro Z28 13.3 8 3.840
Pontiac Firebird 19.2 8 3.845
Fiat X1-9 27.3 4 1.935
Porsche 914-2 26.0 4 2.140
Lotus Europa 30.4 4 1.513
Ford Pantera L 15.8 8 3.170
Ferrari Dino 19.7 6 2.770
Maserati Bora 15.0 8 3.570
Volvo 142E 21.4 4 2.780# Returns columns mpg, cyl, wt for rows with mpg > 25 and wt <= 2
select(filter(mtcars, mpg > 25, wt <= 2), mpg, cyl, wt) mpg cyl wt
Honda Civic 30.4 4 1.615
Toyota Corolla 33.9 4 1.835
Fiat X1-9 27.3 4 1.935
Lotus Europa 30.4 4 1.513dplyr - Pipeline operator and Arrange
As you apply more operations to your dataframe, the nested functions you use can get hard to read.
Luckily,
dplyrincludes the useful pipeline operator%>%(orally referred to as “pipe”, such as in “A pipe B”). Think of it as “and then”.
mpg cyl disp hp drat wt qsec vs am gear carb
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2# Do the same, AND THEN select certain columns
mtcars %>%
filter(mpg > 25, wt <=2) %>%
select(mpg, cyl, wt) mpg cyl wt
Honda Civic 30.4 4 1.615
Toyota Corolla 33.9 4 1.835
Fiat X1-9 27.3 4 1.935
Lotus Europa 30.4 4 1.513########################### arrange ##################################
# Sort the rows in ascending order of wt
mtcars %>%
filter(mpg > 25) %>%
arrange(wt) mpg cyl disp hp drat wt qsec vs am gear carb
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 mpg cyl disp hp drat wt qsec vs am gear carb
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2dplyr - Mutate
mutate()allows you to easily create new variables (as function of other variables)
mtcars %>%
mutate(double_wt = wt*2, # Creates a new variable called double_wt
kms_per_gallon = mpg*1.61, # Creates a new variable called kms_per_gallon
name = row.names(mtcars)) %>% # Creates a new variable with names of the cars
select(name, double_wt, kms_per_gallon) name double_wt kms_per_gallon
Mazda RX4 Mazda RX4 5.240 33.810
Mazda RX4 Wag Mazda RX4 Wag 5.750 33.810
Datsun 710 Datsun 710 4.640 36.708
Hornet 4 Drive Hornet 4 Drive 6.430 34.454
Hornet Sportabout Hornet Sportabout 6.880 30.107
Valiant Valiant 6.920 29.141
Duster 360 Duster 360 7.140 23.023
Merc 240D Merc 240D 6.380 39.284
Merc 230 Merc 230 6.300 36.708
Merc 280 Merc 280 6.880 30.912
Merc 280C Merc 280C 6.880 28.658
Merc 450SE Merc 450SE 8.140 26.404
Merc 450SL Merc 450SL 7.460 27.853
Merc 450SLC Merc 450SLC 7.560 24.472
Cadillac Fleetwood Cadillac Fleetwood 10.500 16.744
Lincoln Continental Lincoln Continental 10.848 16.744
Chrysler Imperial Chrysler Imperial 10.690 23.667
Fiat 128 Fiat 128 4.400 52.164
Honda Civic Honda Civic 3.230 48.944
Toyota Corolla Toyota Corolla 3.670 54.579
Toyota Corona Toyota Corona 4.930 34.615
Dodge Challenger Dodge Challenger 7.040 24.955
AMC Javelin AMC Javelin 6.870 24.472
Camaro Z28 Camaro Z28 7.680 21.413
Pontiac Firebird Pontiac Firebird 7.690 30.912
Fiat X1-9 Fiat X1-9 3.870 43.953
Porsche 914-2 Porsche 914-2 4.280 41.860
Lotus Europa Lotus Europa 3.026 48.944
Ford Pantera L Ford Pantera L 6.340 25.438
Ferrari Dino Ferrari Dino 5.540 31.717
Maserati Bora Maserati Bora 7.140 24.150
Volvo 142E Volvo 142E 5.560 34.454dplyr - Why use the pipeline operator?
The pipeline operator makes it much easier to read what is going on. Note that the following two pieces of code achieve the same thing.
mpg wt cyl
Fiat 128 32.4 2.200 4
Porsche 914-2 26.0 2.140 4
Fiat X1-9 27.3 1.935 4
Toyota Corolla 33.9 1.835 4
Honda Civic 30.4 1.615 4
Lotus Europa 30.4 1.513 4 mpg wt cyl
Fiat 128 32.4 2.200 4
Porsche 914-2 26.0 2.140 4
Fiat X1-9 27.3 1.935 4
Toyota Corolla 33.9 1.835 4
Honda Civic 30.4 1.615 4
Lotus Europa 30.4 1.513 4You need some concentration to get what the first code is doing! In contrast, you can easily interpret the second code as:
take the
mtcarsdataset, and thenfilter for
mpg > 25, and thenselect the columns
mpg,wt,cyl, and thenarrange them by descending
weight
dplyr - summarise and group_by
summarise() will give you summary statistics
mean_mpg num_cars sd_wt
1 20.09062 32 0.9784574group_by() is a very powerful function that will group rows into different categories. You can then use summarise() to find summary statistics of each group
mtcars %>%
group_by(cyl) %>% # Group cars by how many cylinders they have
summarise(avg_mpg = mean(mpg)) # Find the average mpg for each group (cyl)# A tibble: 3 × 2
cyl avg_mpg
<dbl> <dbl>
1 4 26.7
2 6 19.7
3 8 15.1Common summary statistics include:
mean()the averagesd()the standard deviationn()the count
This can also be used to group by multiple features
dplyr Exercise 1
Using the in-built dataset quakes:
Find all the rows with magnitude (
mag) greater or equal to 5.5Display only the columns
depth,magandstations
depth mag stations
1 139 6.1 94
2 50 6.0 83
3 42 5.7 76
4 56 5.7 106
5 127 6.4 122
6 205 5.6 98
7 216 5.7 90
8 577 5.7 104
9 562 5.6 80
10 48 5.7 123
11 535 5.7 112
12 64 5.9 118
13 75 5.6 79
14 546 5.7 99
15 417 5.6 129
16 153 5.6 87
17 93 5.6 94
18 183 5.6 109
19 627 5.9 119
20 40 5.7 78
21 242 6.0 132
22 589 5.6 115
23 107 5.6 121
24 165 6.0 119dplyr Exercise 2
Using the in-built data set mtcars:
Create a new variable which is the product of
mpgtimeswt.Find the mean and median of this new variable, but within each possible value of
cyl. Do you notice something?
dplyr - Why use dplyr?
dplyr is generally easier to read and write than base R. The following codes do the same thing
dplyr:
Base R:
In particular, group_by and summarise are easy to write with dplyr. They are significantly harder to write in base R:
dplyr:
Base R:
avg_weight <- aggregate(mtcars$wt,
by = list(cylinders = mtcars$cyl,
gears = mtcars$gear),
FUN = mean)
names(avg_weight)[3] <- "Average_weight"
sd_weight <- aggregate(mtcars$wt,
by = list(cylinders = mtcars$cyl,
gears = mtcars$gear),
FUN = sd)
names(sd_weight)[3] <- "std_dev_weight"
new_df <- merge(avg_weight, sd_weight, by = c("cylinders", "gears"))Or to count the number of observations within given intervals:
dplyr:
sample_data %>%
mutate(ints = cut(height, breaks= c(140,150, 160, 170, 180, 190), right = F)) %>%
group_by(ints) %>%
summarise(n = n())# A tibble: 5 × 2
ints n
<fct> <int>
1 [140,150) 3
2 [150,160) 70
3 [160,170) 89
4 [170,180) 52
5 [180,190) 12Base R:
# put the data in 5 categories
res <- hist(height, plot=FALSE, breaks = c(140,150,160,170,180,190), right = F)
x <- as.table(res$counts) # create the table
nn <- as.character(res$breaks)
# add names to categories: this step is not straightforward
dimnames(x) <- list(paste(nn[-length(nn)], nn[-1],sep="-"))
x140-150 150-160 160-170 170-180 180-190
3 70 89 52 12 dplyr - comments and more information
Cheat sheet for
dplyr- https://github.com/rstudio/cheatsheets/blob/main/data-transformation.pdfOnline tutorials
- the dplyr website
- the data transformation chapter in “R for Data Science”.
To go further in your
Rlearning journey, the aforementioned R for Data Science textbook is one of the most popular and useful resources out there.
Note: you may have noticed on some of the previous slides (e.g., slide 25) that dplyr was outputting something called a tibble. These are a more modern version of data frames, designed to work better within the tidyverse ecosystem.
While there are interesting if subtle differences between tibbles and data frames, they should not concern you much at this stage; you can use and manipulate them in almost the same way.
Homework
Homework Exercise 1
Using the iris dataset:
- Find all the rows with
Sepal.Widthgreater than \(2.7\) and then sort bySepal.Length - For all the
setosaflowers, find the ratio betweenSepal.LengthandSepal.Width, call this new variableratioand display only columnsratioandSpecies - Find the average
ratiobetweenSepal.LengthandSepal.Widthfor each species of flower
Homework Exercise 2
Import the dataset sample_data.csv:
- create a new column called
BMI, which is calculated by \[ \frac{\text{Weight}}{\text{Height}(m)^2} \] - calculate the average
BMIfor each of the 6 categories ofcooked_fruit_vegfor FEMALE individuals only
Homework Exercise 3
The built-in dataset ChickWeight records the weights of chicks (in the variable weight), as well as the type of diet (\(1, 2, 3\) or \(4\) in the variable Diet).
Provide summary statistics for the weight in diet groups \(1\) to \(4\). Can you say for sure that a given group of chicks is the heaviest?
Home work Exercise 4
In Ed, the dataset motor.df is already loaded in the script on your right. This dataset contains some information about a large number of vehicle insurance claims, by policy. Perform the following tasks:
- Display the first 10 rows of the dataset
motor.df, and have a look at the variables names. - Use the package
dplyrto create a summary table (or “tibble”) which should have two columns:
- The first reports the occurrence date of the accident, or
AccidentDay, but converted to the ‘date’ format. Hint: use functionas.Date(). - The second is named
ncand reports, byAccidentDay, the total number of claims on each day.
- Use the table created in Q2 as well as function
plot()to produce a scatterplot of the daily numbers of CTP claims (y axis) from the year 2006 to the year 2015 (x axis is time).
Home work Exercise 5
In Ed, the dataset school.data is already loaded in the script on your right. This dataset contains some information about every school in Australia (primary and secondary), in the year 2019.
Use the package dplyr to create a summary table (or “tibble”) called teaching.staff. This table should have three columns:
- The first reports the
School.Sector. - The second is named
nband reports, bySchool.Sector, the total number of schools (in all Australia). - The third is named
mean.studentsand reports, bySchool.Sector, the mean of a new variable which you must create, and which is the ratio of variableTotal.Enrolmentsdivided byFull.Time.Teaching.Staff. Hint: you may need to use argumentna.rmin functionmean().
Hint: the expected result should look like this: