library(tidyverse)
PCarIns <- read_csv("PrivateCarIns1975-Data.csv")
PCarIns <- PCarIns %>% filter(Numb.Claims>0) # remove the 5 categories with no claims
str(PCarIns)spc_tbl_ [123 × 7] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ Pol.Age : num [1:123] 1 1 1 1 1 1 1 1 1 1 ...
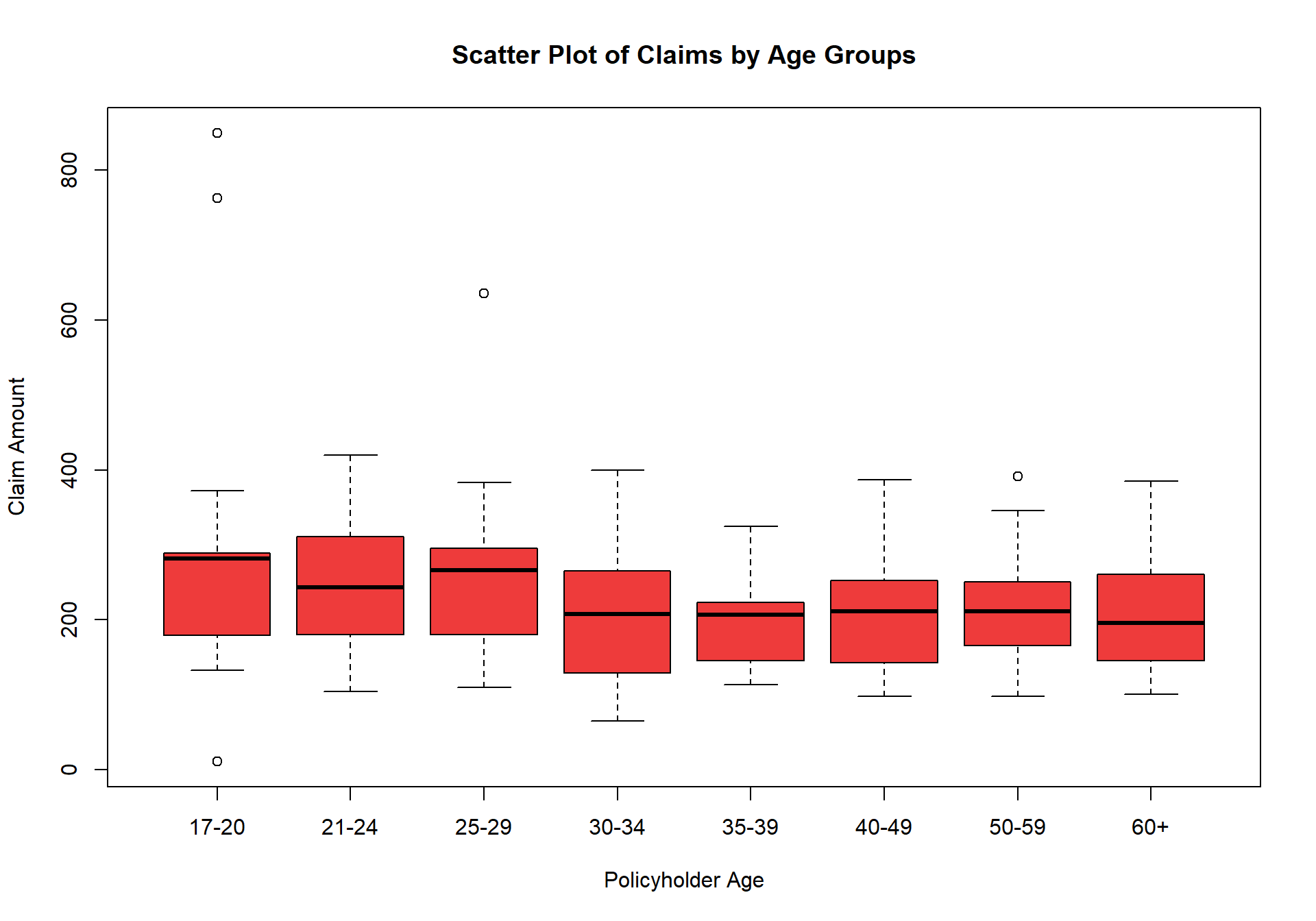
$ Cpol.Age : chr [1:123] "17-20" "17-20" "17-20" "17-20" ...
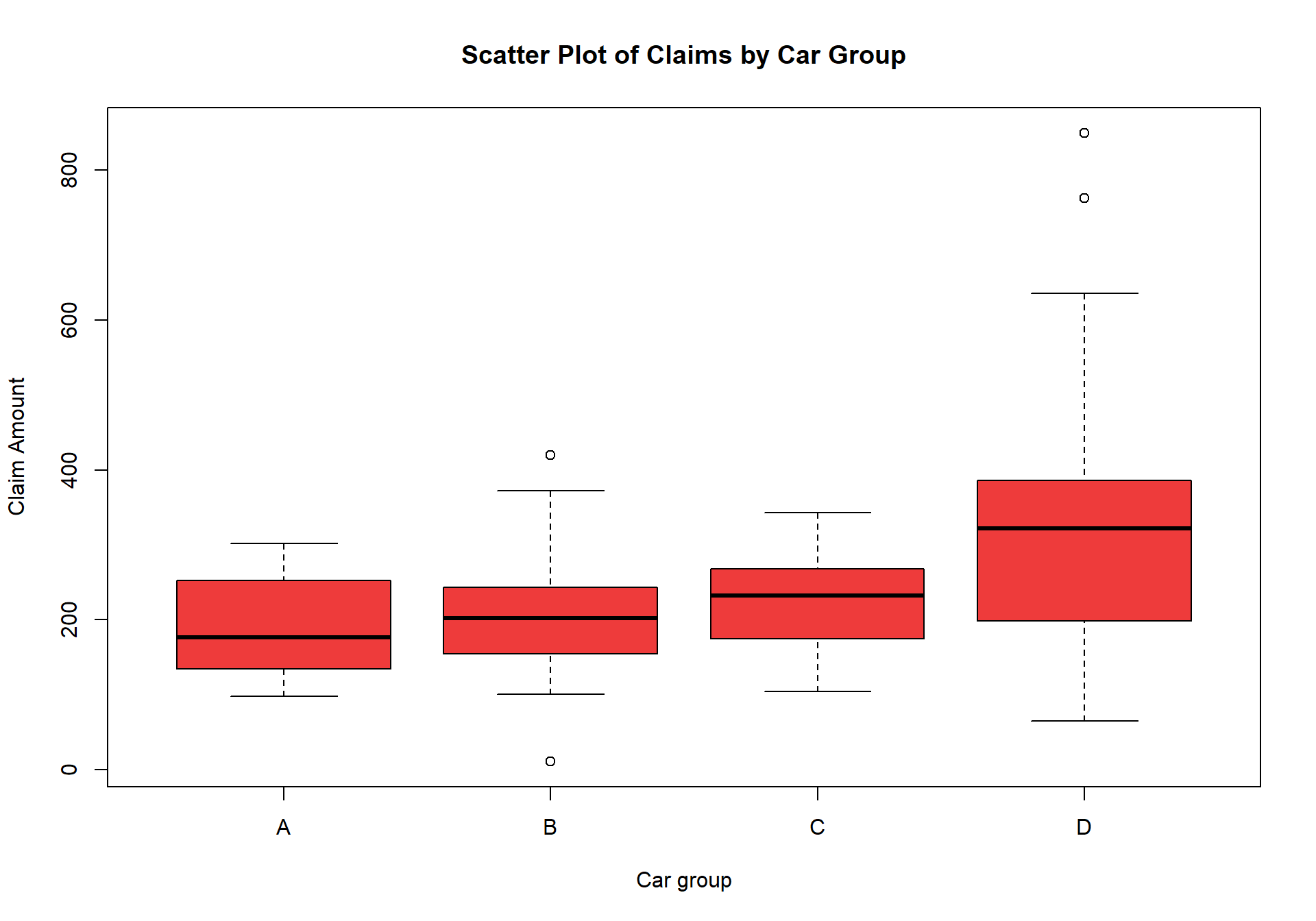
$ Car.Group : chr [1:123] "A" "A" "A" "A" ...
$ Veh.Age : num [1:123] 1 2 3 4 1 2 3 4 1 2 ...
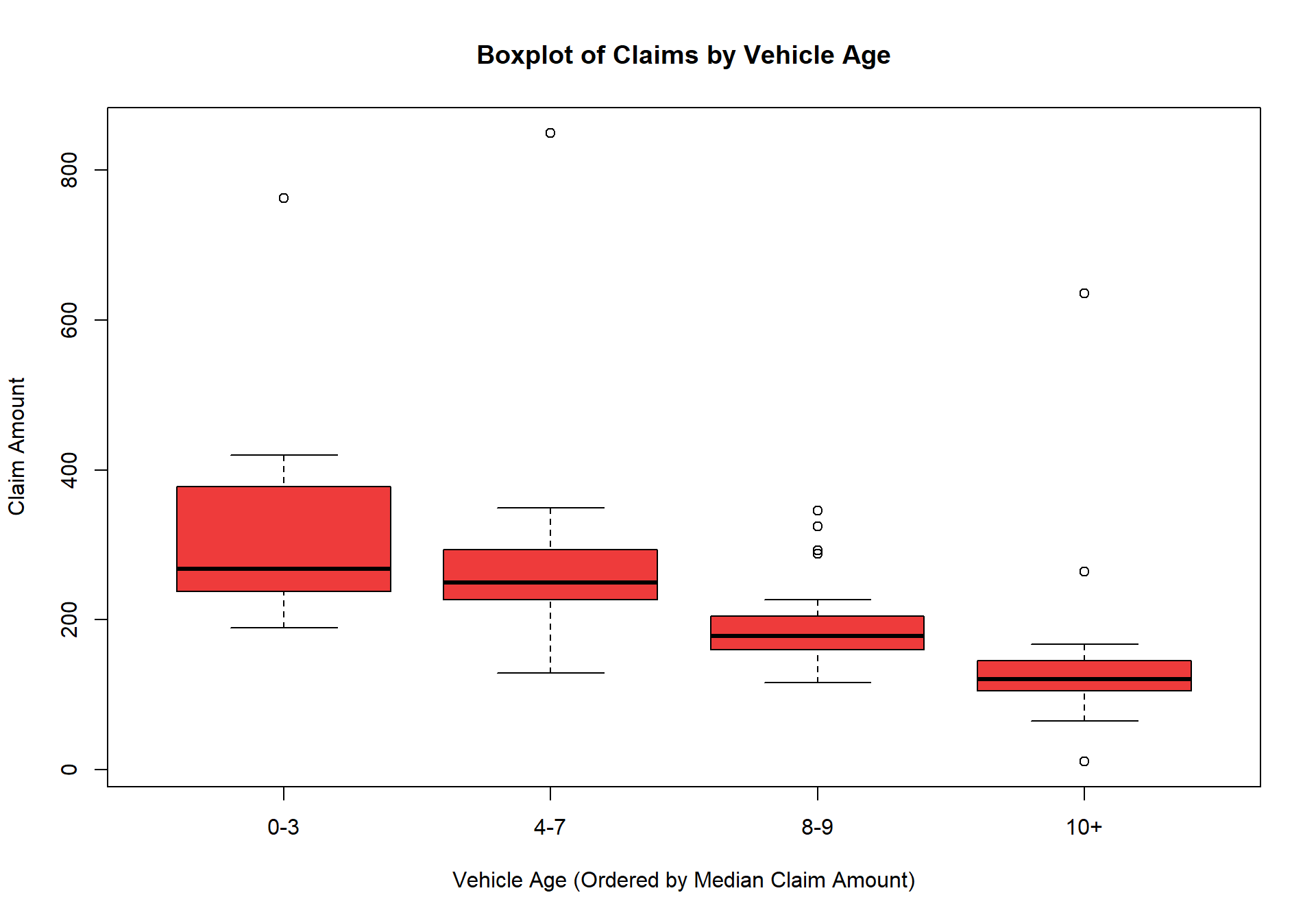
$ Cveh.Age : chr [1:123] "0-3" "4-7" "8-9" "10+" ...
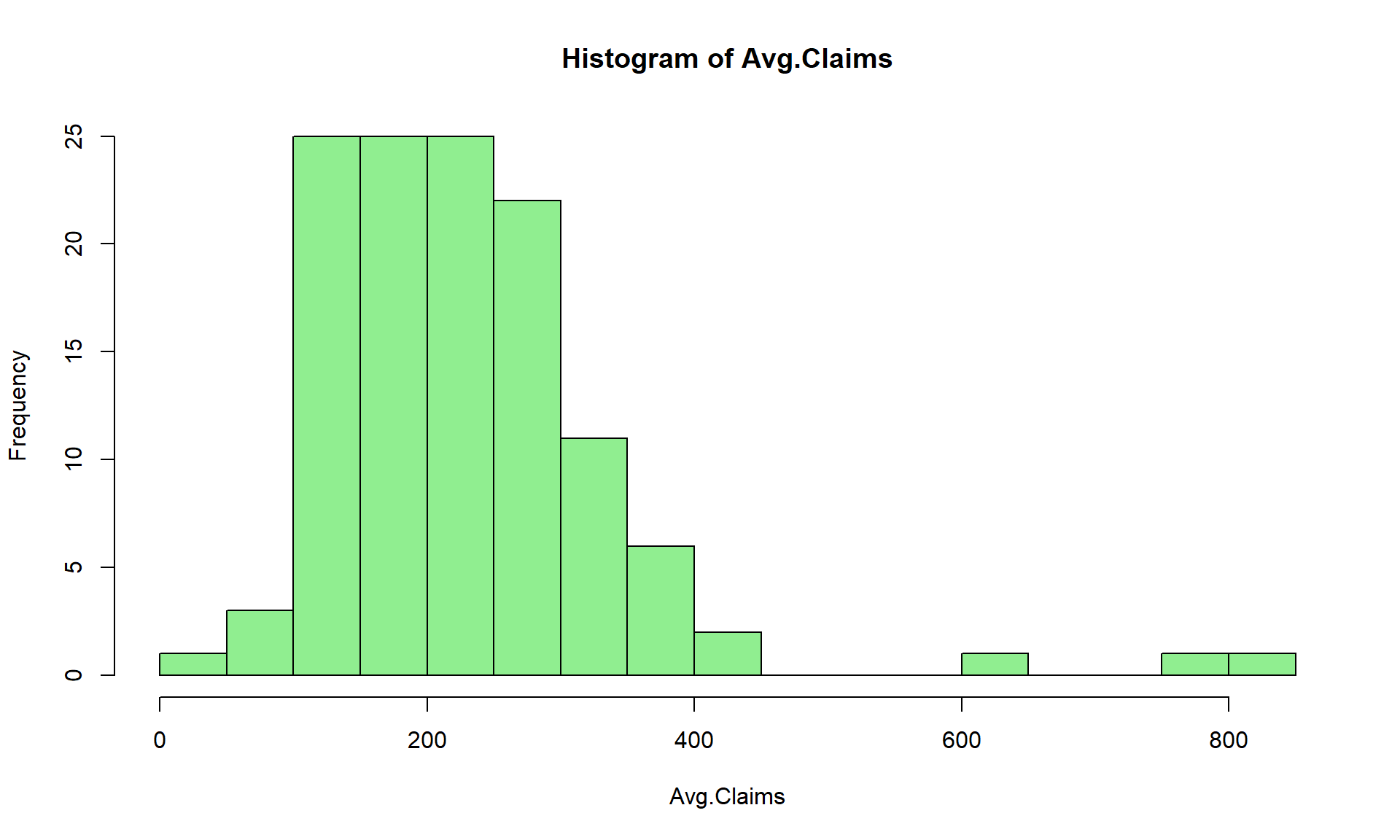
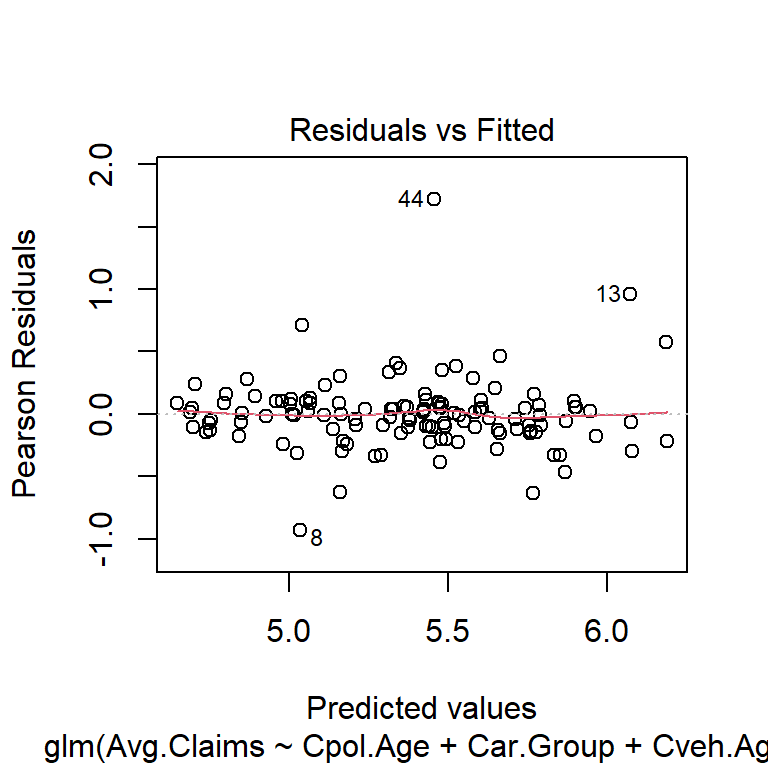
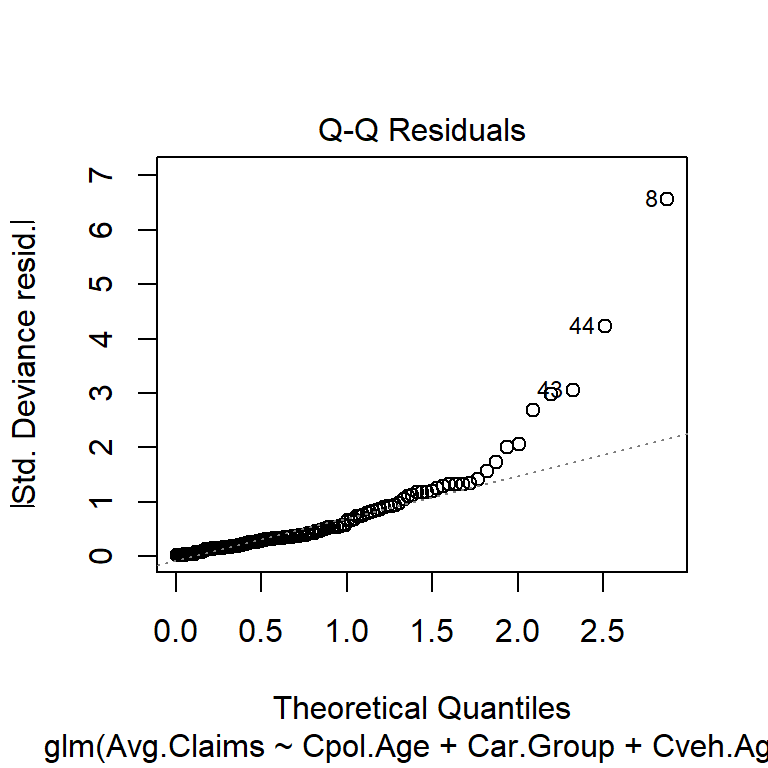
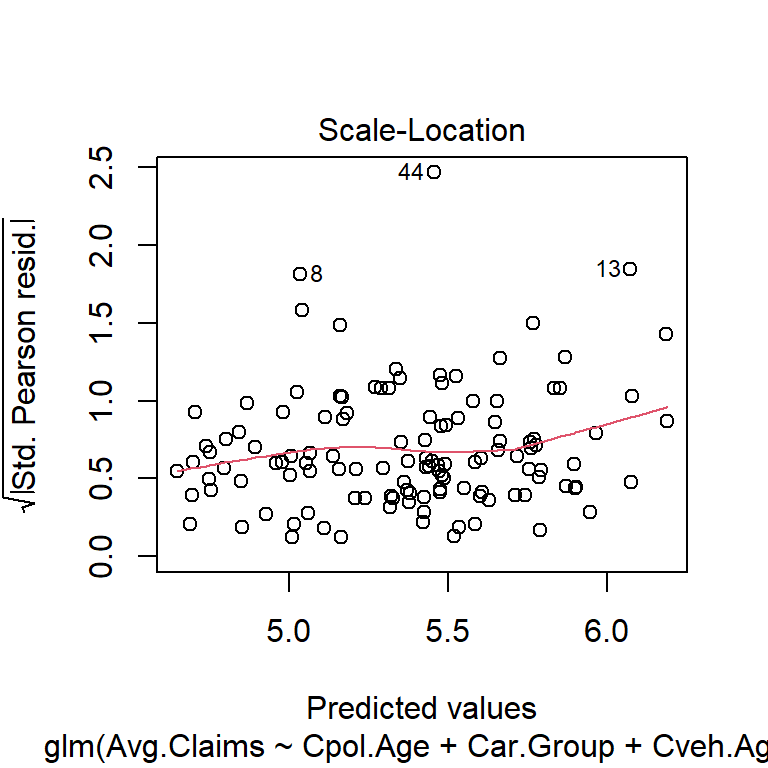
$ Avg.Claims : num [1:123] 289 282 133 160 372 249 288 11 189 288 ...
$ Numb.Claims: num [1:123] 8 8 4 1 10 28 1 1 9 13 ...
- attr(*, "spec")=
.. cols(
.. Pol.Age = col_double(),
.. Cpol.Age = col_character(),
.. Car.Group = col_character(),
.. Veh.Age = col_double(),
.. Cveh.Age = col_character(),
.. Avg.Claims = col_double(),
.. Numb.Claims = col_double()
.. )
- attr(*, "problems")=<externalptr>