ggplot(raw_data)
By the end of this topic, students should be able to
draw and edit curves and plots with ggplot2;
present and summarise statistical data with visualisation tools in gglot2;
know where to look for to further learn R
ggplot2 is a package in R that offers a wide range of visualisation tools.
The language of ggplot2 is always the same:
It is a natural way of creating plots.
It allows advanced visualisation in a simple way.
The basic function is ggplot()
ggplot(raw_data)
An empty plot is created.
A dataset is supplied, but with no further instruction on how the data should be presented, hence nothing is displayed.
Importantly, the data set supplied (here ‘raw_data’) must be a data frame.
A geom is a function that returns a layer of visualisation.
There are many different geom in ggplot2, to visualise data in different ways:
geom_point - scatter graphgeom_line - line graphgeom_boxplot - box plotgeom_histogram - histogramgeom_abline - adds a straight line, specified by intercept and slopegeom_smooth - adds a line/curve of best fitNow, let’s use geom_point to create a scatter graph.
ggplot(raw_data)+geom_point(aes(x = height, y = weight))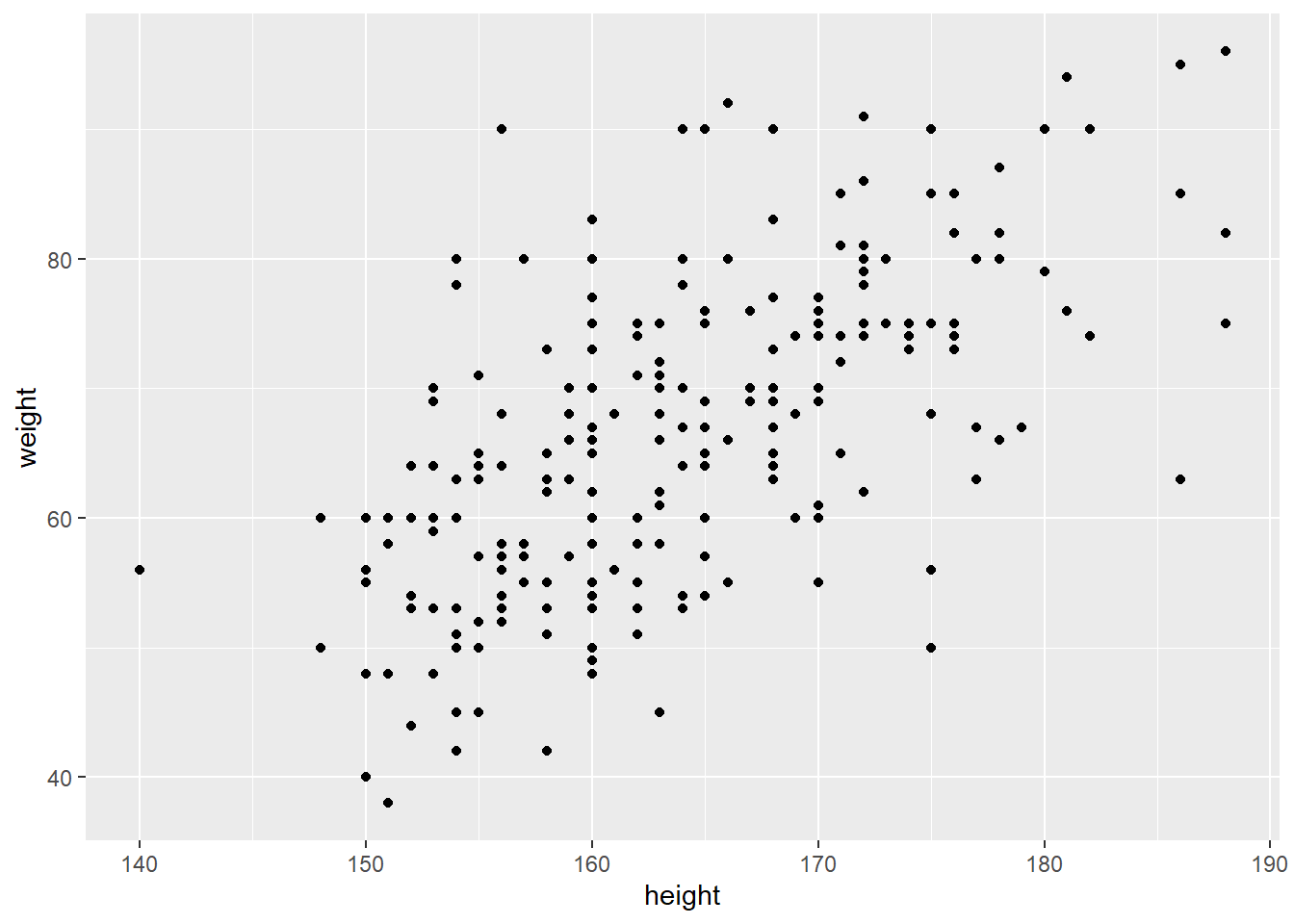
ggplot() function.aes() stands for ‘aesthetic’. Different geoms have different required aesthetics but these are generally features of the visualisation that should be determined by the data.x and y coordinates to be determined by the data, hence we use aes(x = height, y = weight)geom_point is actually “inheriting” the data argument raw_data from ggplot(raw_data). It would be equally valid to write:ggplot() + geom_point(raw_data, aes(x = height, y = weight))We will see more on this and how it can be used later.
Using sample_data.csv, create a boxplot for the variable weight by the variable meat.
raw_data<-read.csv("sample_data.csv",header=TRUE)
ggplot(raw_data)+geom_boxplot(aes(meat,weight))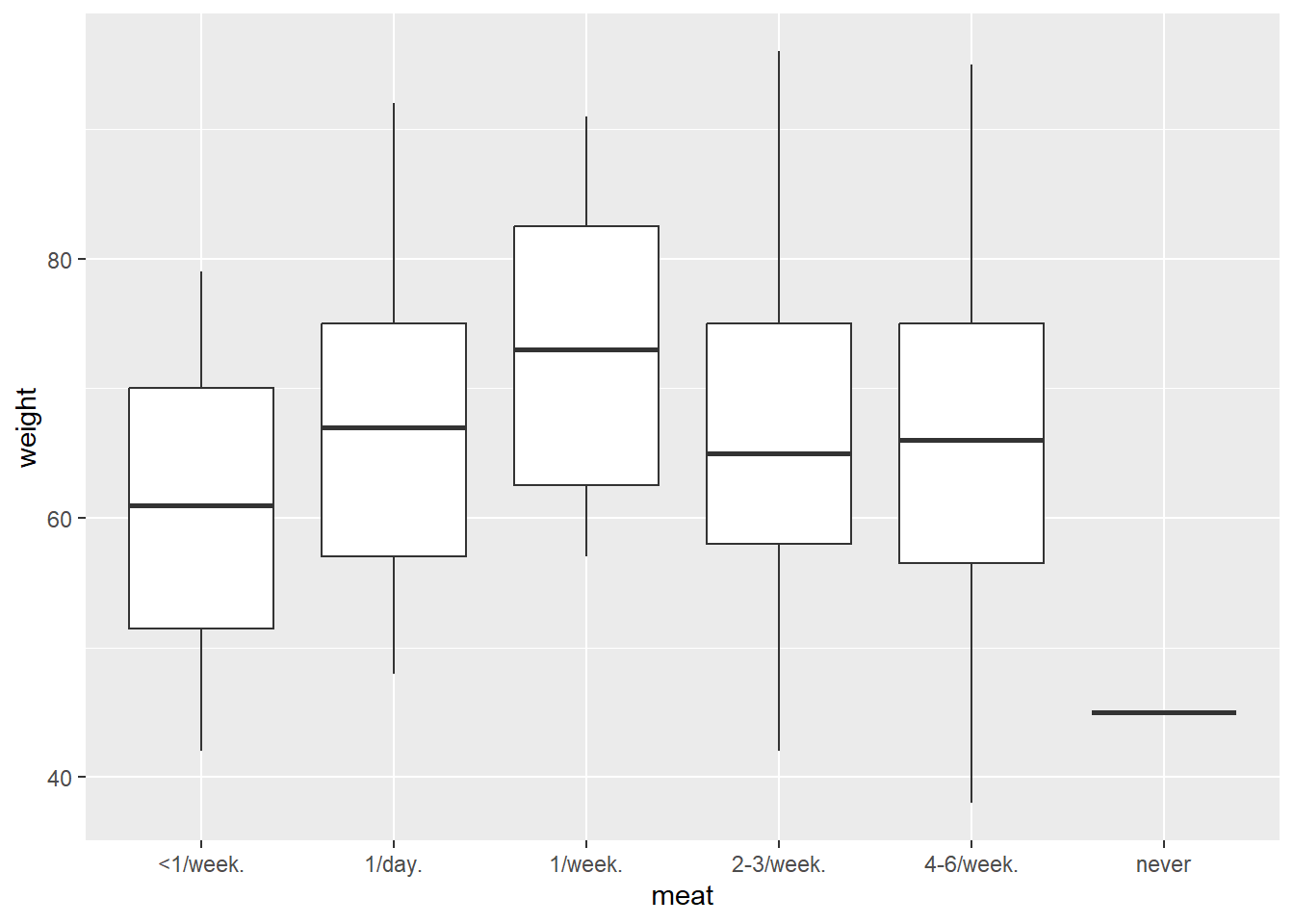
ggplot(raw_data)+
geom_point(aes(height, weight))+ # draw points
geom_abline(intercept = -70, slope = 0.8) # add line (note this is not the best fit, just a line with arbitrary values)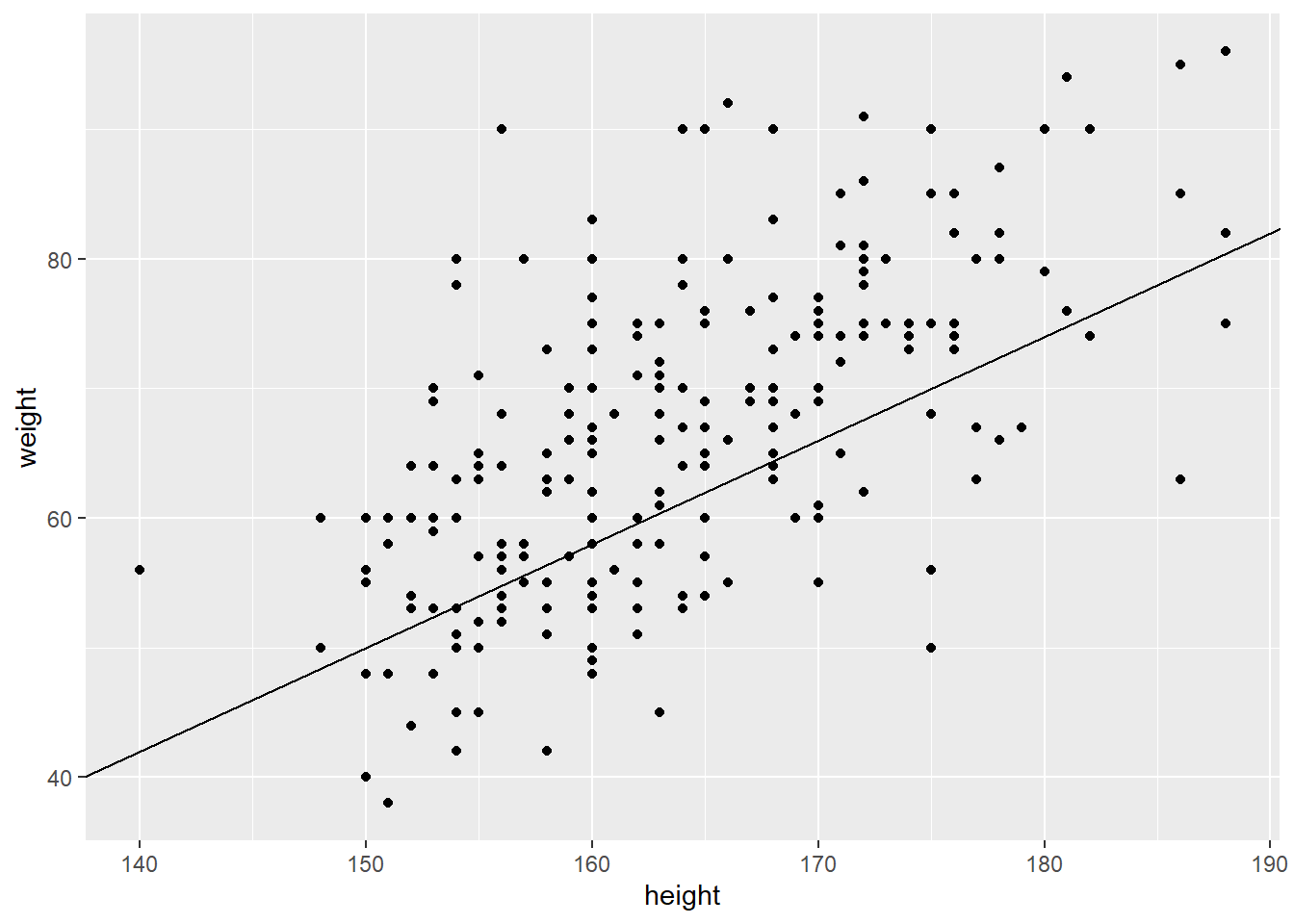
One can also create the line with the geom_smooth() function.
ggplot(raw_data, aes(height, weight))+
geom_point()+
geom_smooth(method = "lm", se = TRUE)+ # this adds the best linear fit to the data
labs(x = "Height", y = "Weight",
title ="Line Chart of Height against Weight",
subtitle = "A simple illustration",
caption = "This graph shows the relationship between height and weight")
## `geom_smooth()` using formula = 'y ~ x'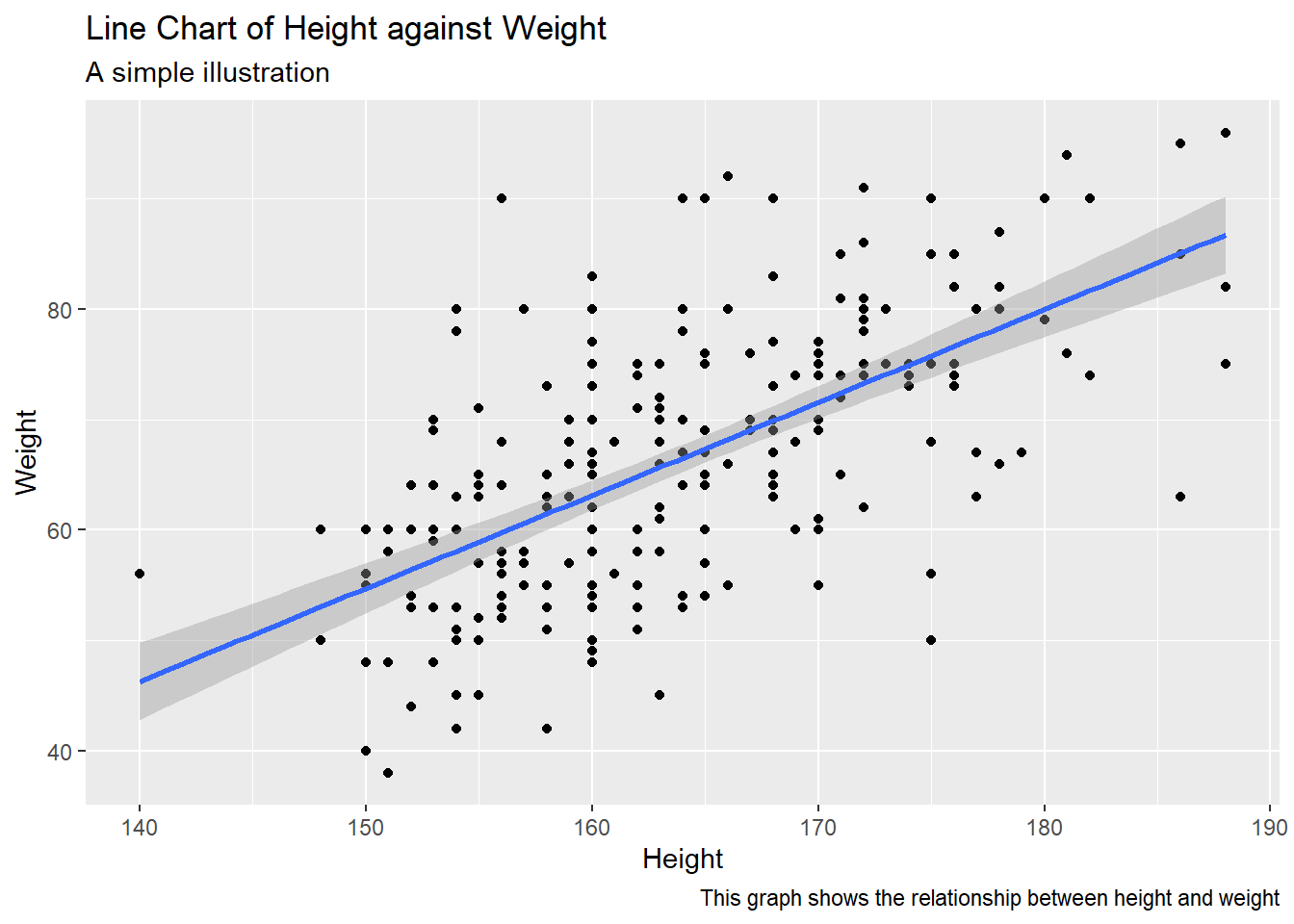
Notes:
se=TRUE displays the confidence interval on the fitted line (stay tuned for the maths of this in ACTL2131, a second year course)ggplot. In this case, raw_data and aes(height, weight) were inherited by both geom_point and geom_smooth, and in fact any other further geoms would also inherit them. This significantly reduces repetition when it is appropriate.labs is used to customise any fixed label, i.e. x and y axes, the main title, legend titles, etc.Use the iris dataset. Create a histogram for the variable Petal.Length. Add appropriate labels (x-axis, y-axis) and a title to the plot.
Note that one should adjust the binwidth.
ggplot(iris)+
geom_histogram(aes(Petal.Length), binwidth=0.5)+
labs(x = "Weight", y = "Number of observations",
title ="Histogram for the variable Petal.Length")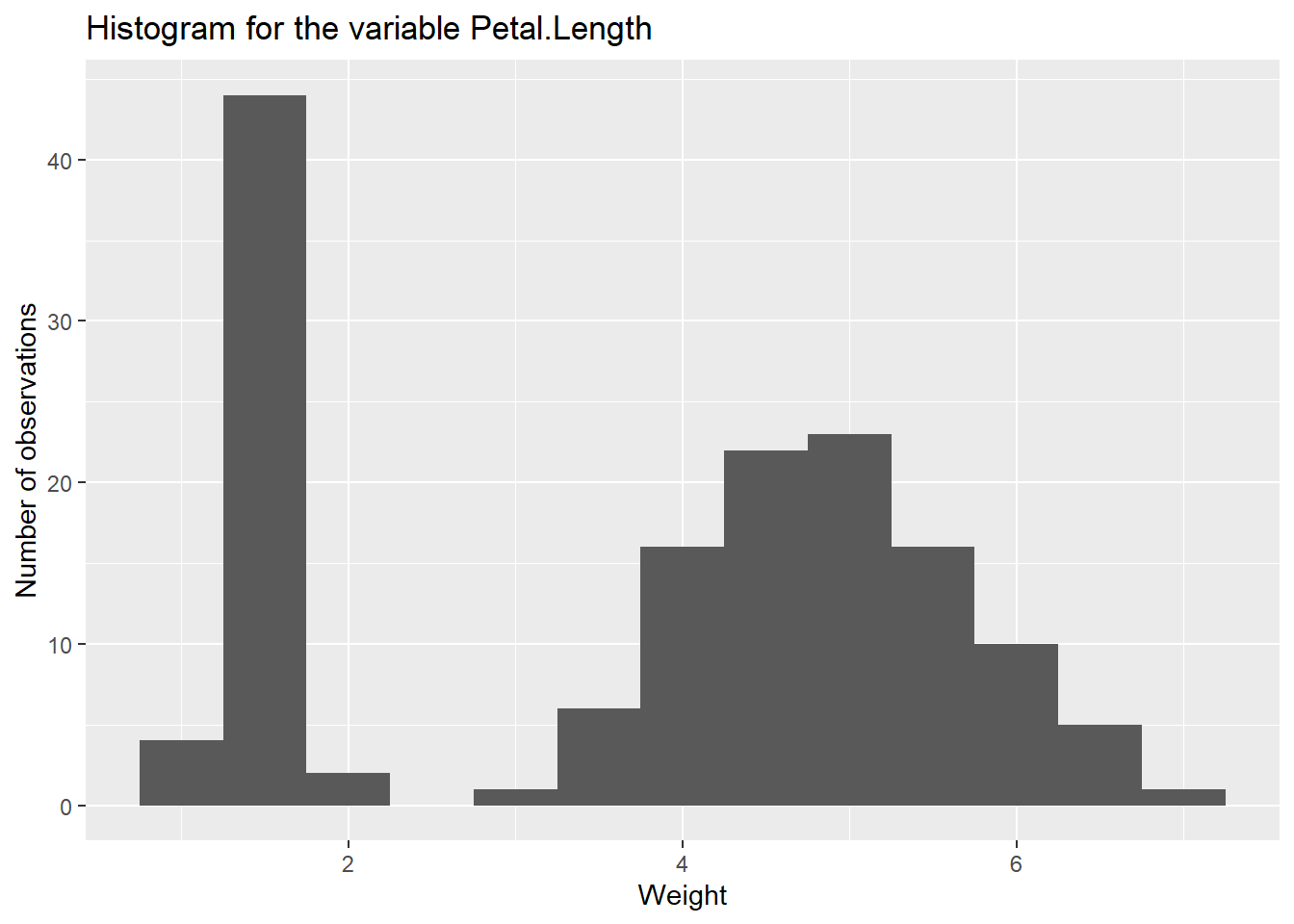
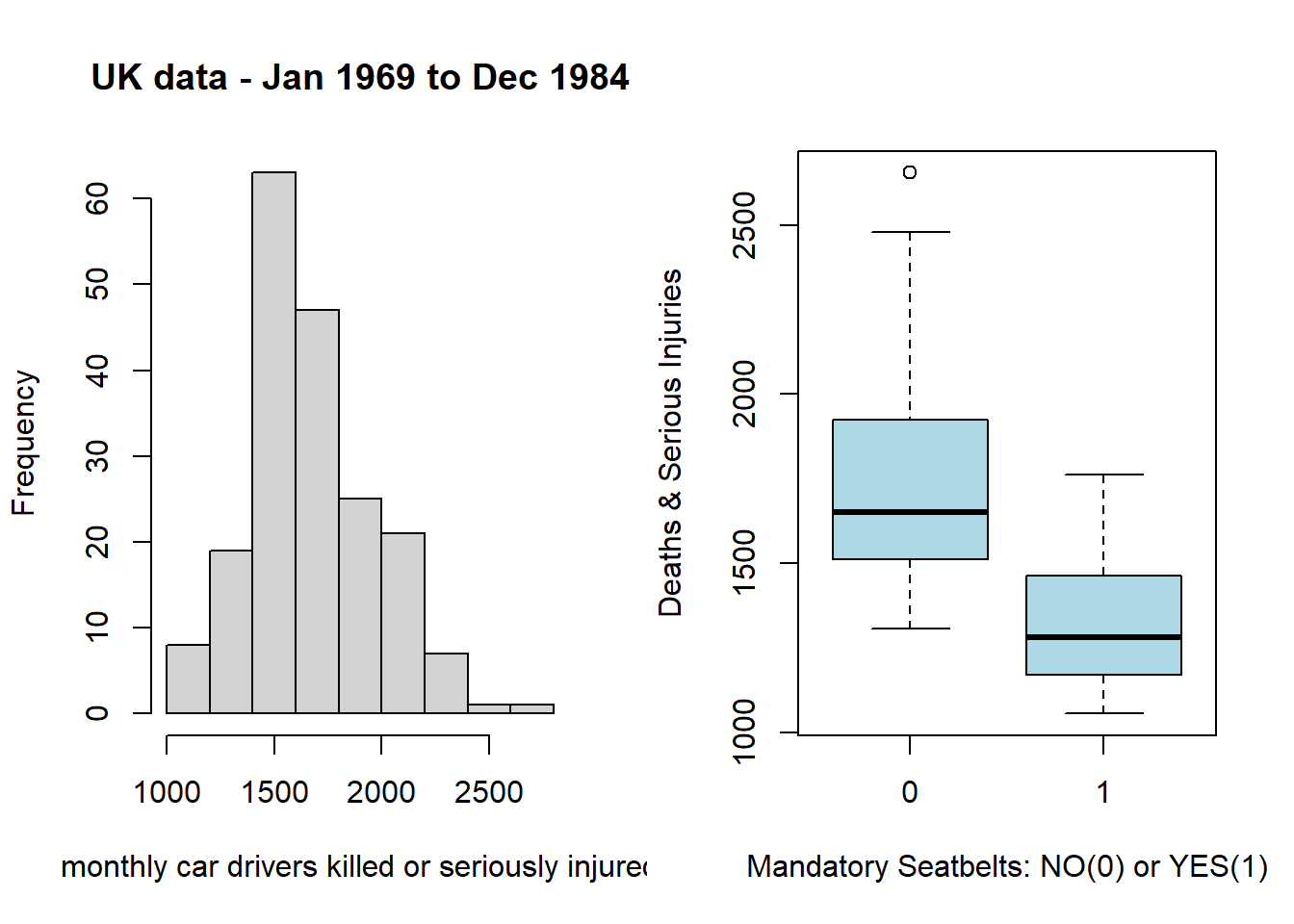
Use the Seatbelts dataset to recreate the following two graphs from last week’s lab in ggplot: a histogram of the drivers column, and a boxplot of drivers by law. Do not worry about the colouring or theme for now, we will discuss that later.
data <- as.data.frame(Seatbelts)
ggplot(data) +
geom_histogram(aes(drivers), bins = 10) +
labs(x = "monthly car drivers killed or seriously injured",
title = "Histogram of UK - Jan 1969 to Dec 1984")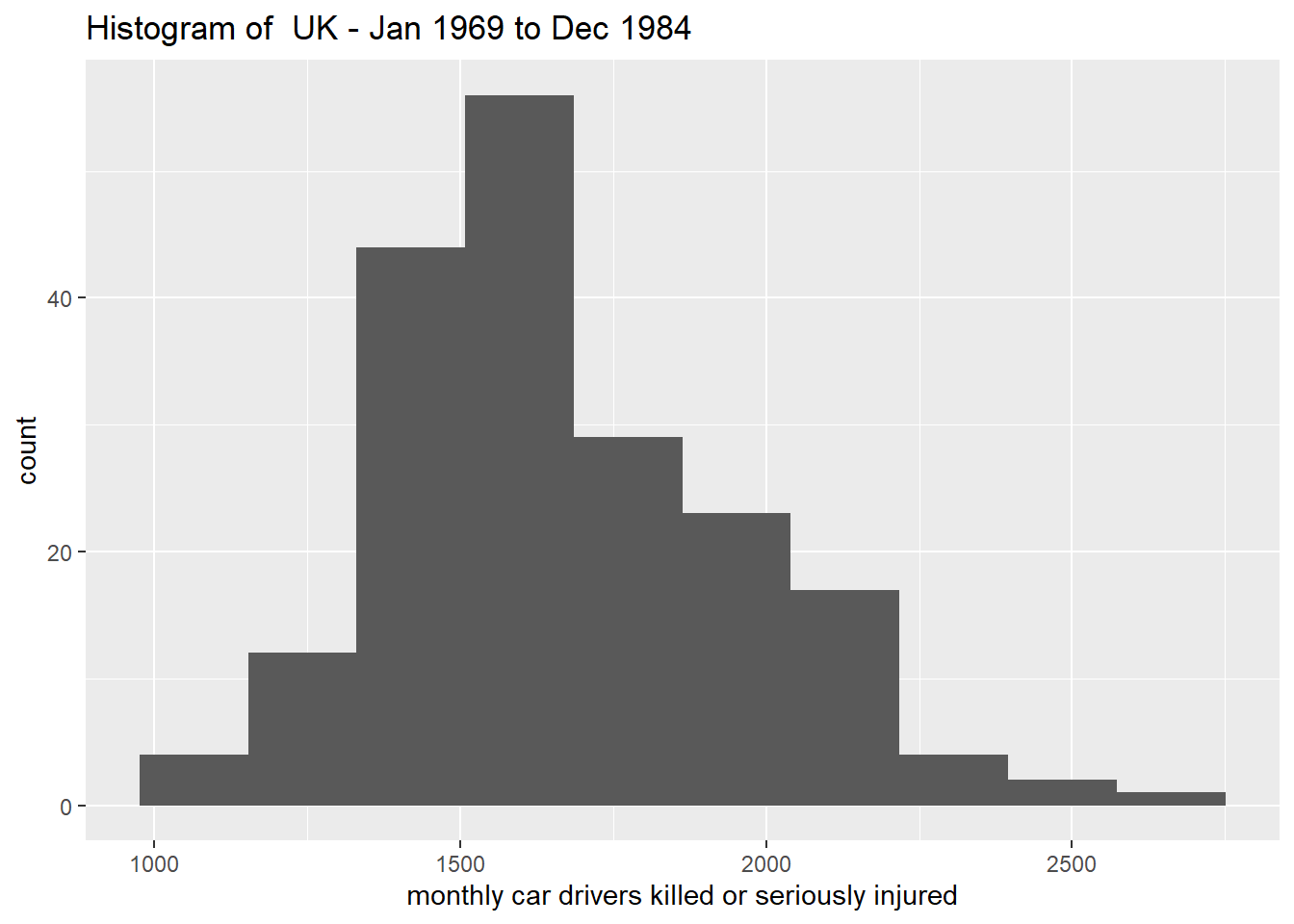
# convert variable 'law' in a factor
data$law <- factor(data$law, labels = c("0", "1"))
ggplot(data) +
geom_boxplot(aes(x = law, y = drivers)) +
labs(x = "Mandatory Seatbelts: NO(0) or YES(1)",
y = "Deaths & Serious Injuries")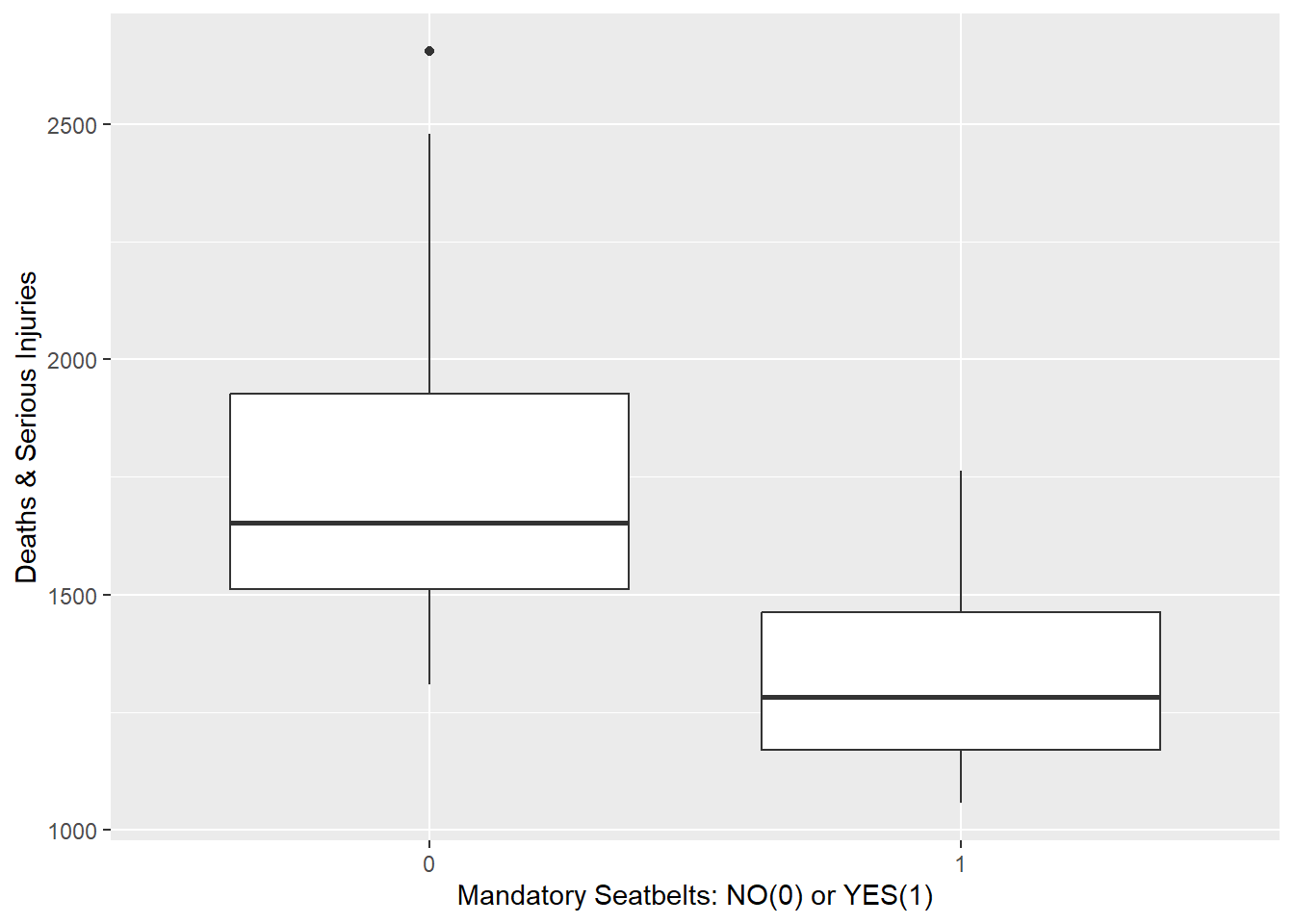
Another type of visual layer you can add with ggplot2 is a stat. For example, you can use the stat_ecdf() to visualise the empirical distribution function of a continuous variable.
ggplot(raw_data, aes(height)) +
stat_ecdf() + labs(y = 'Empirical CDF')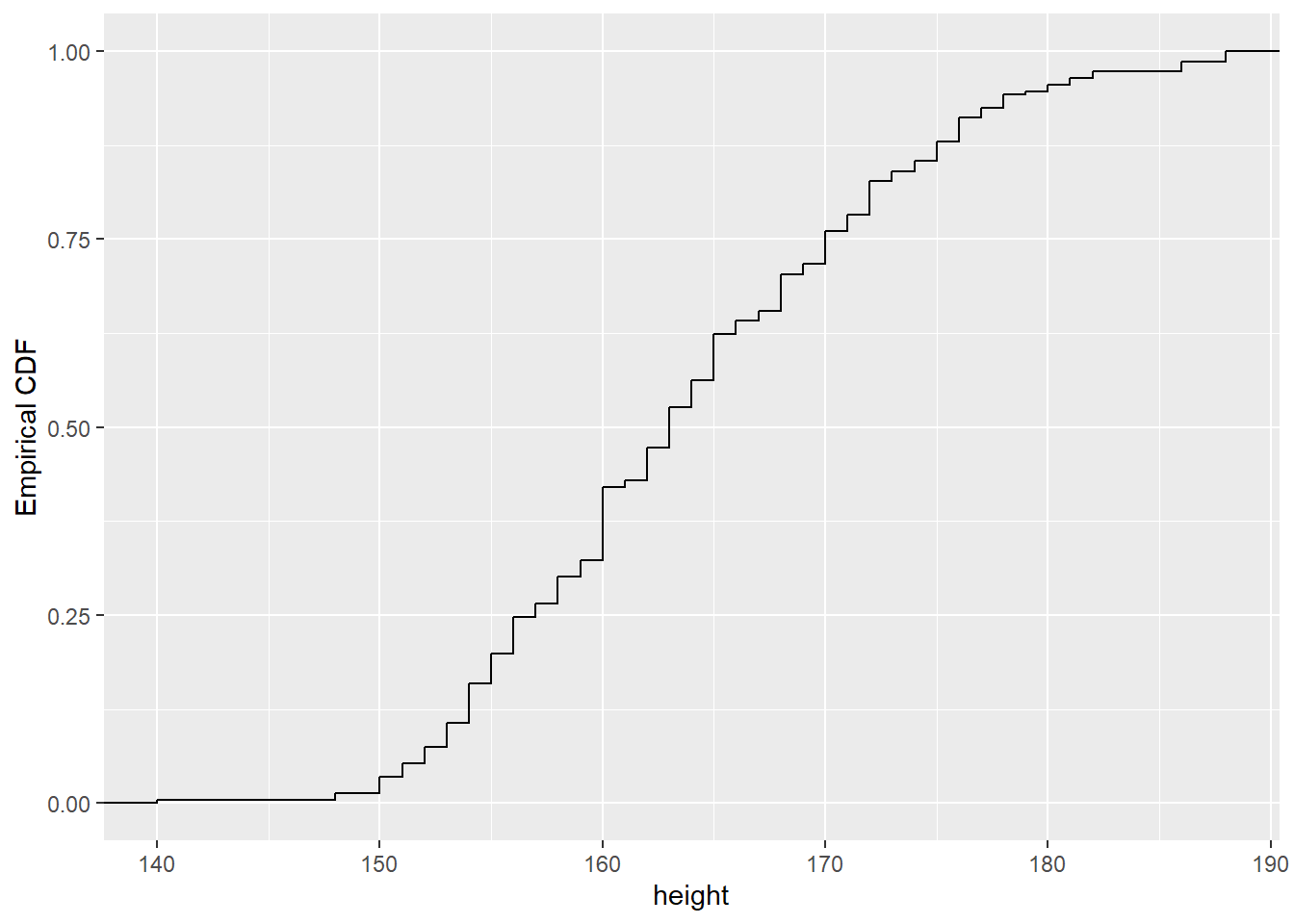
stat and a geom are required for any visual layer!geom_point, it actually is by default selecting the geom point and the stat identitystat_ecdf is defaulting to the stat ecdf and the geom stepstats are used when ggplot2 needs to calculate a new variable to plot
stat_function can be used to directly plot a function, for example a normal distribution density function as below
ggplot(data = data.frame(x = c(-3, 3)), aes(x)) +
stat_function(fun = dnorm, n = 100, args = list(mean = 0, sd = 1))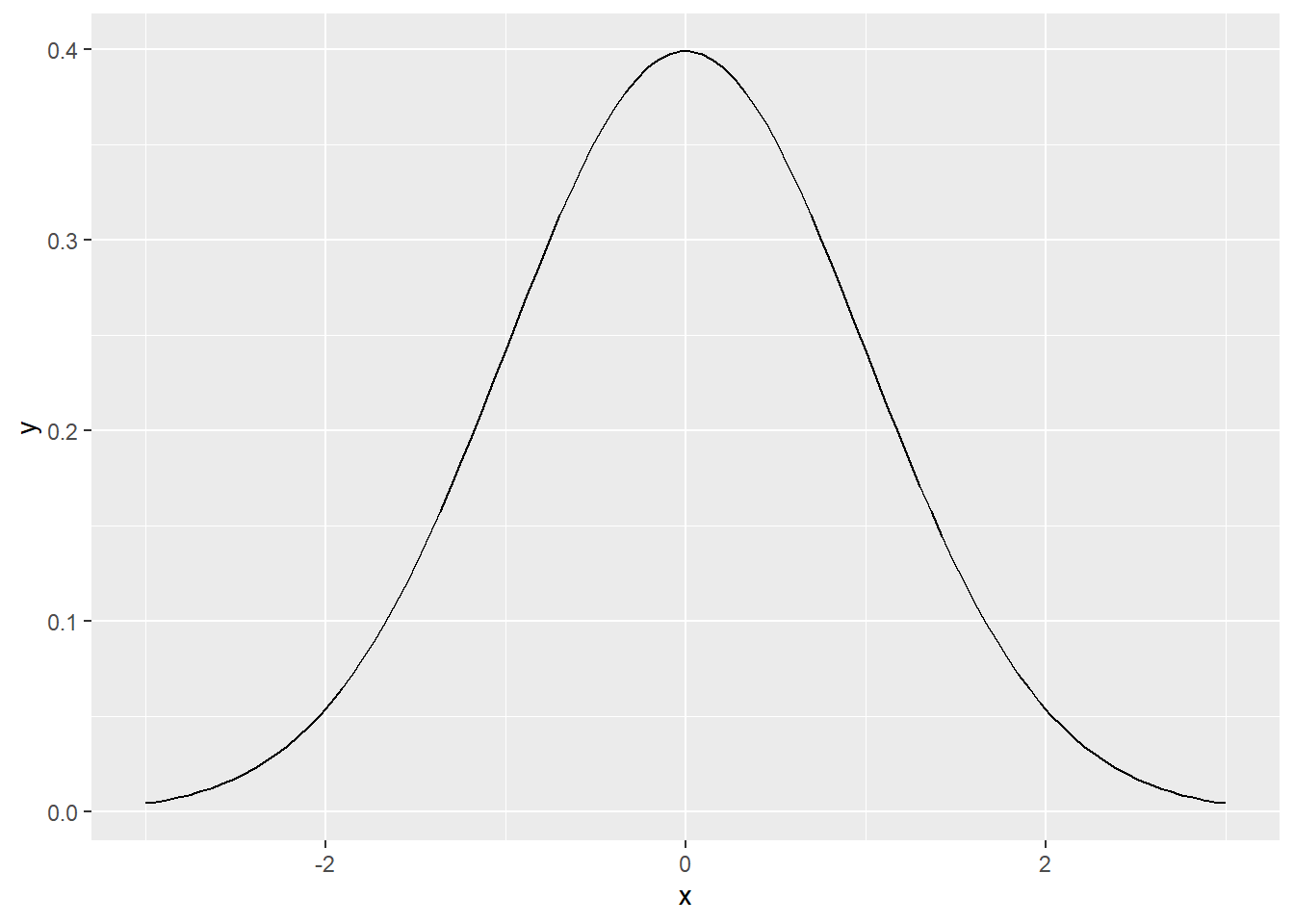
Use stat_function to plot:
A lognormal(0,1) cumulative distribution function on the interval(0,10)
The function:
\[y=x^2+2x+4-e^{1-x^2}\] on the interval(-1,1)
ggplot(data = data.frame(x = c(0, 10)), aes(x)) +
stat_function(fun = plnorm, n = 100, args = list(mean = 0, sd = 1))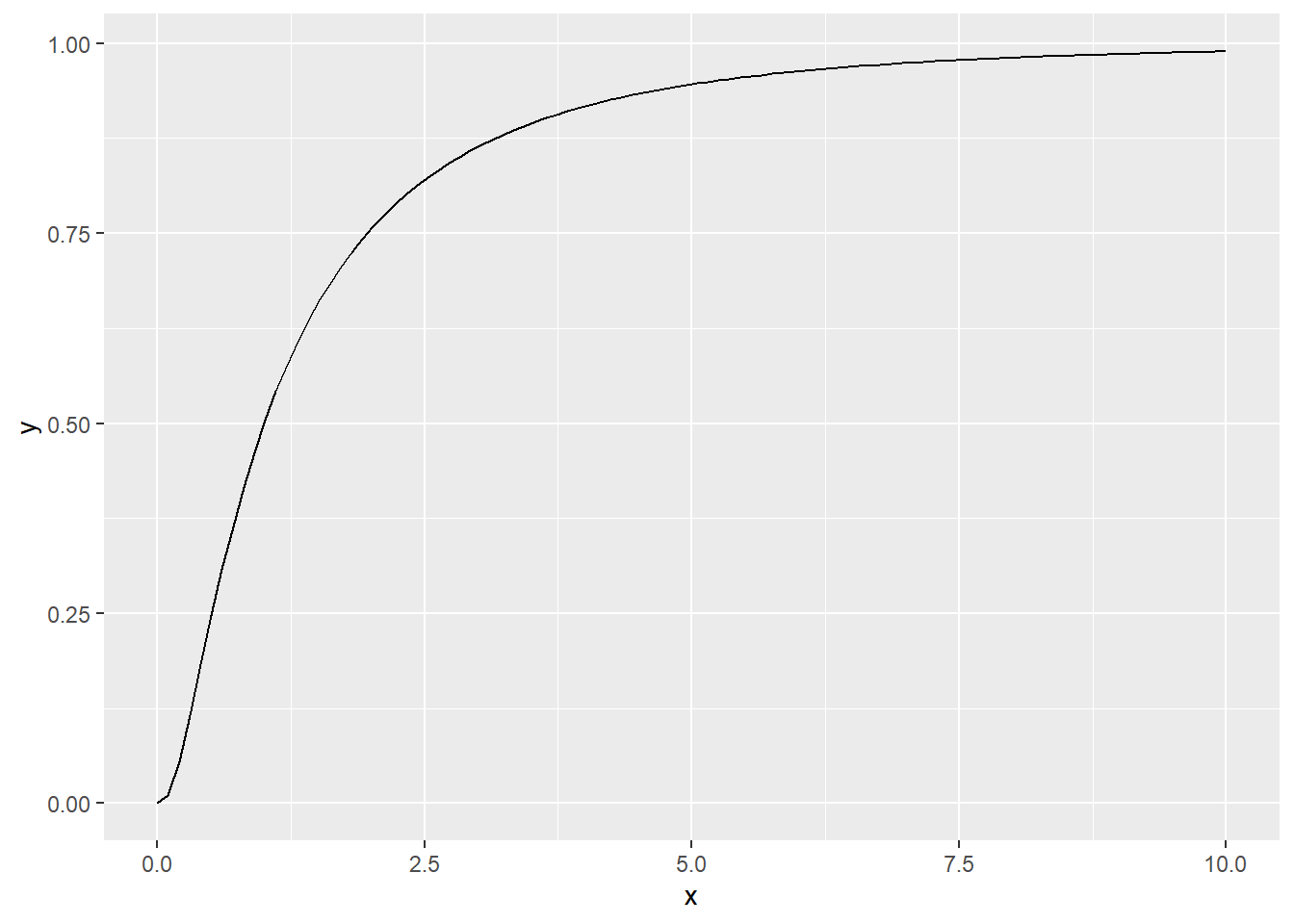
f = function(x) {
return(x^2+2*x+4-exp(1-x^2))
}
ggplot(data = data.frame(x = c(-1, 1)), aes(x)) +
stat_function(fun = f, n = 100)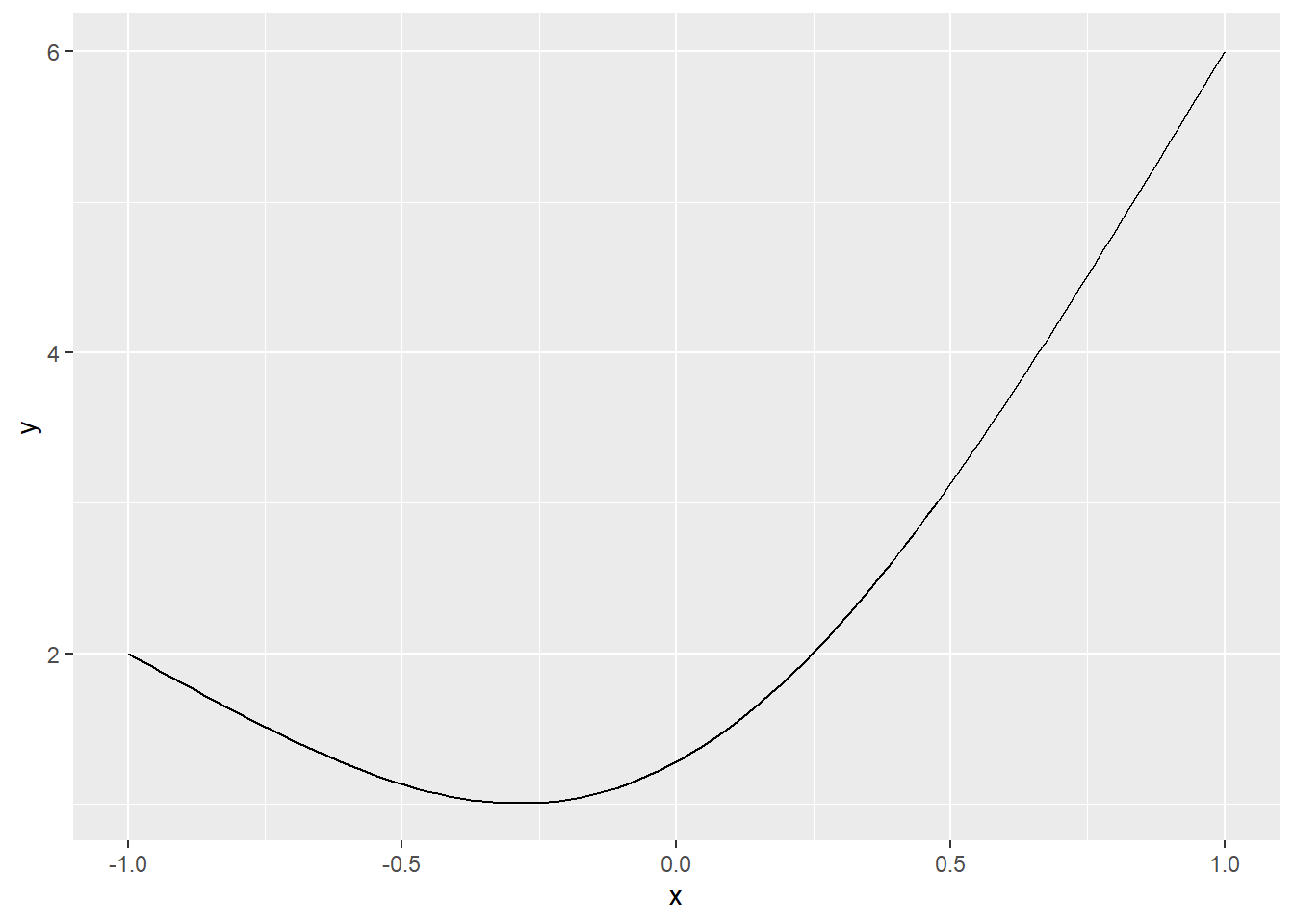
ggplot2 offers a range of built-in themes to improve the presentation of your plots, by changing their overall aspect
ggplot(raw_data,aes(height))+stat_ecdf()+theme_bw()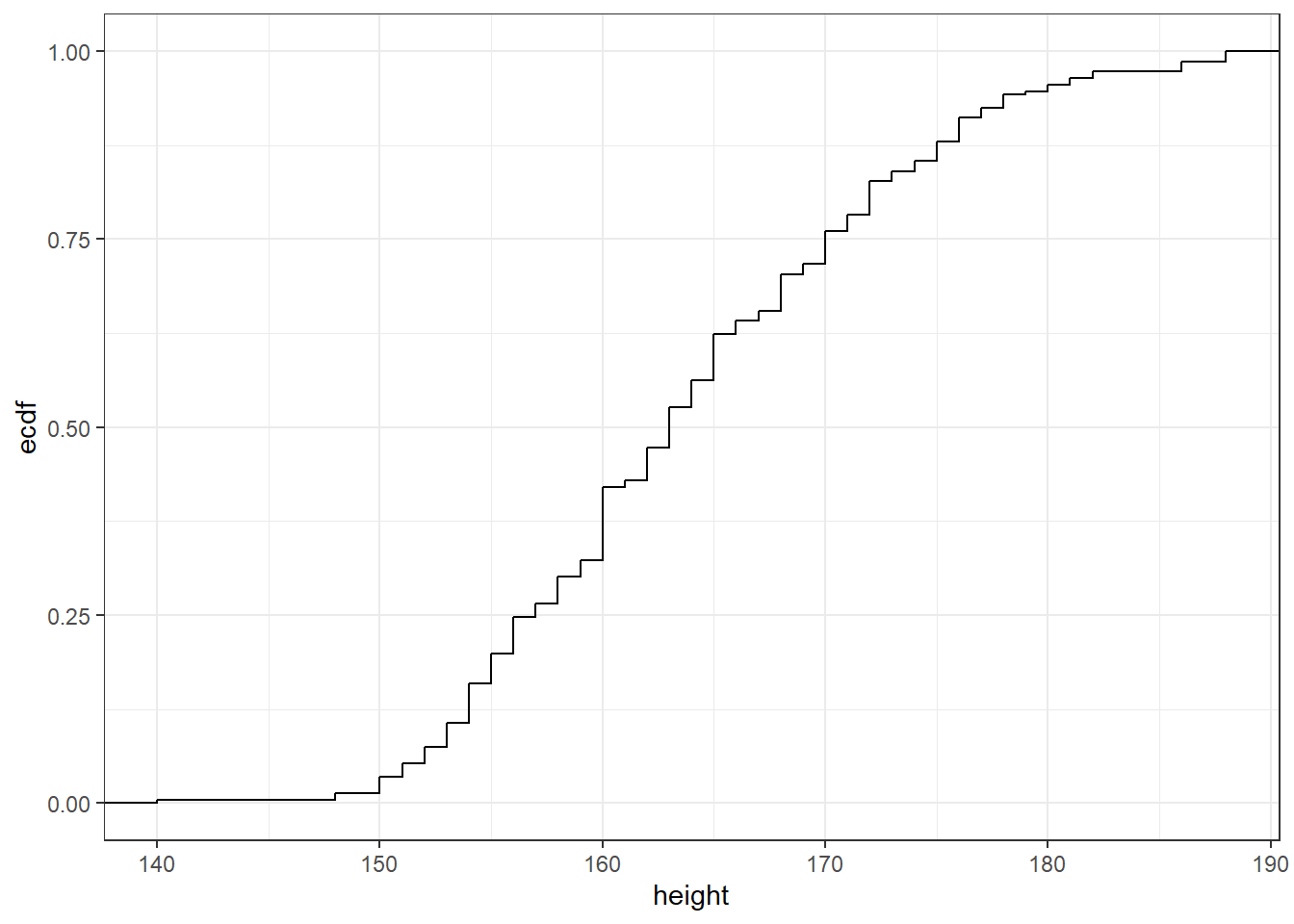
ggplot(raw_data,aes(height))+stat_ecdf()+theme_classic()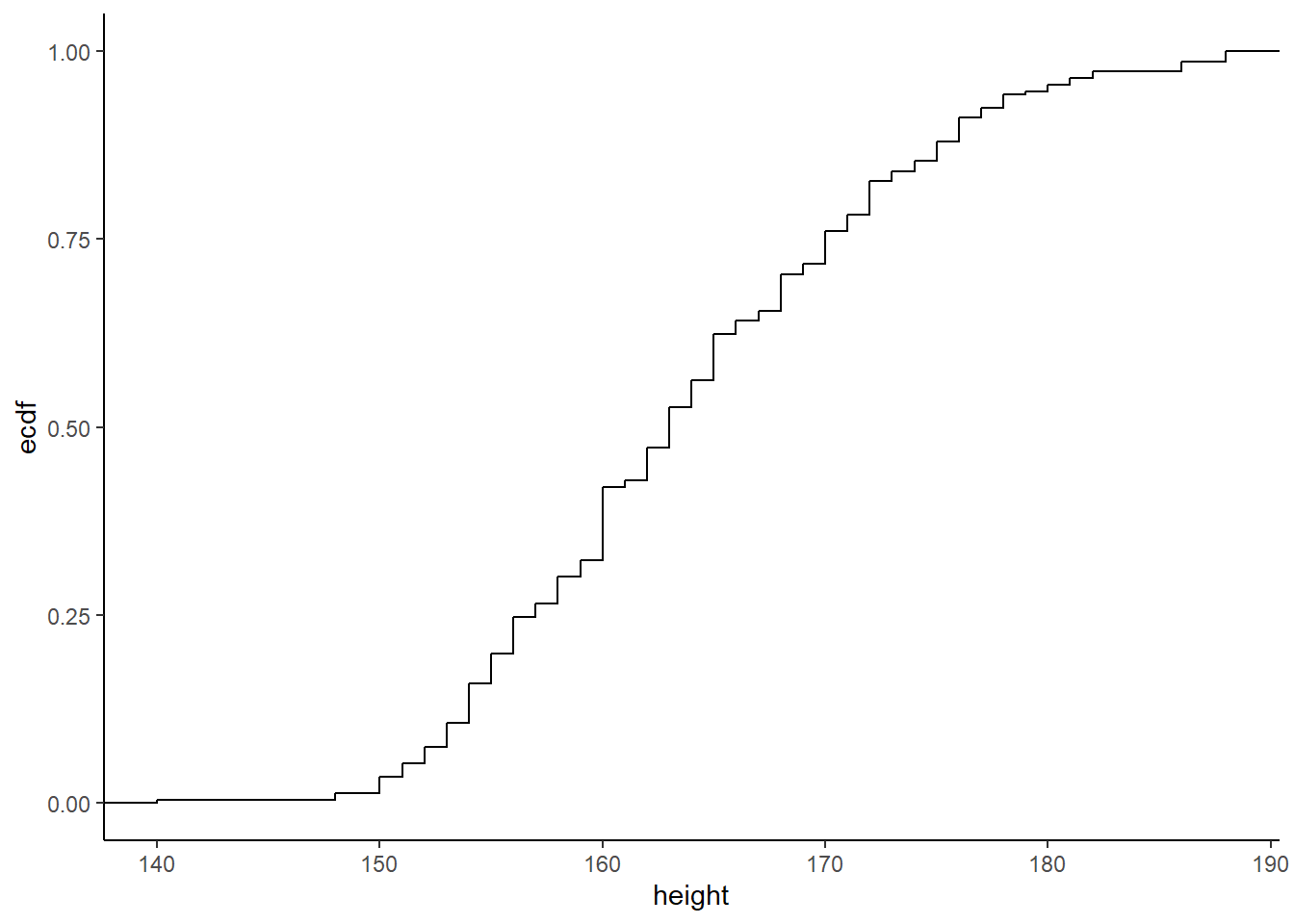
ggplot(raw_data,aes(height))+stat_ecdf()+theme_dark()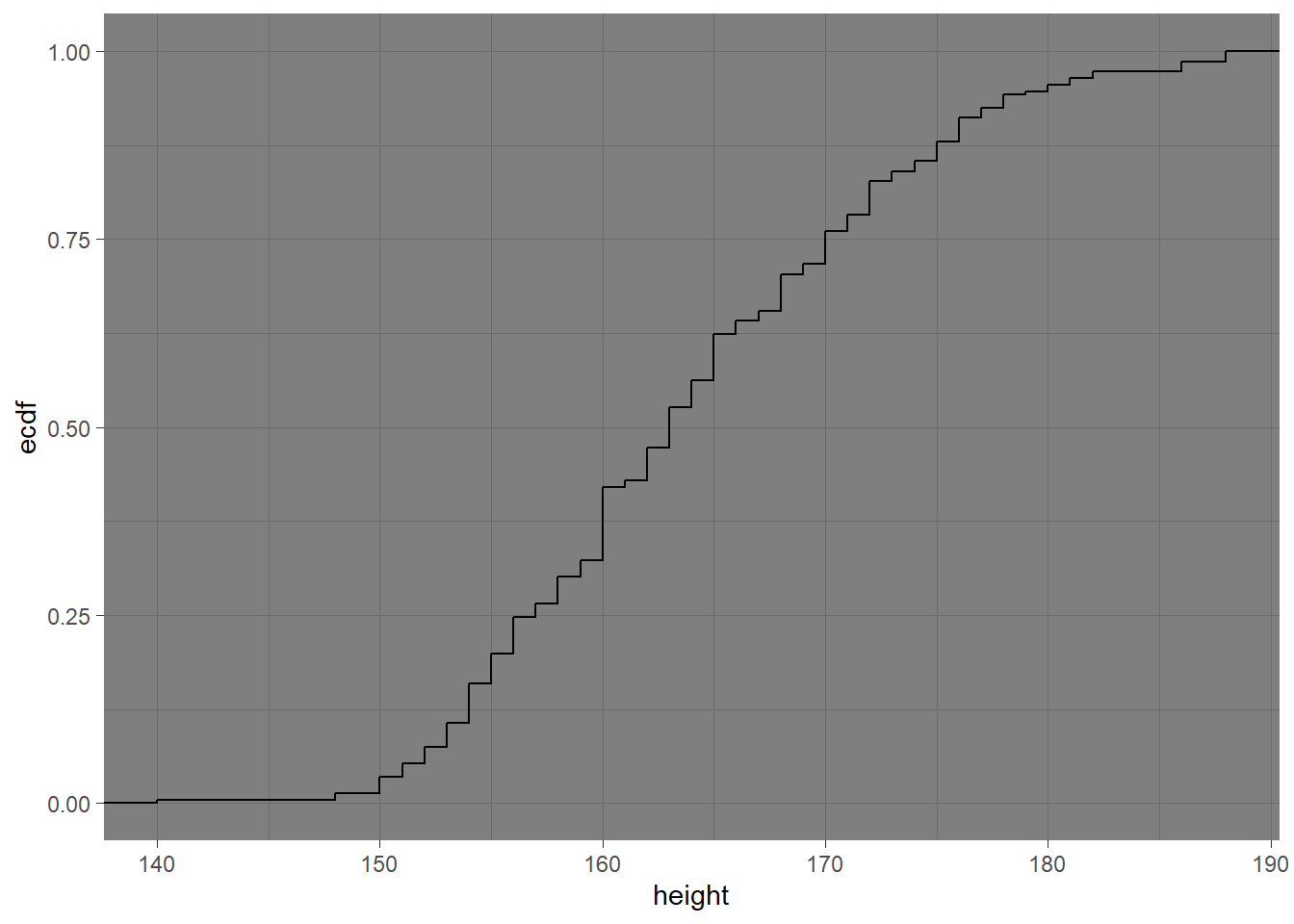
An important part of visualisation is categorising different observations, for example by colour, shape, size or transparency. ggplot provides convenient syntax for this.
For example, say we want to produce a plot like this:
We start with a plain ggplot. Dataset midwest (also from ggplot2 package) contains demographic information of midwest counties (such as total population, area, state, population density, etc) in the USA.
ggplot(midwest, aes(x=area, y=poptotal)) +
geom_point() +
xlim(c(0, 0.1)) + # plot inside a range that suit us
ylim(c(0, 500000)) +
theme(text = element_text(size=20)) #make axis bigger
## Warning: Removed 15 rows containing missing values or values outside the scale range
## (`geom_point()`).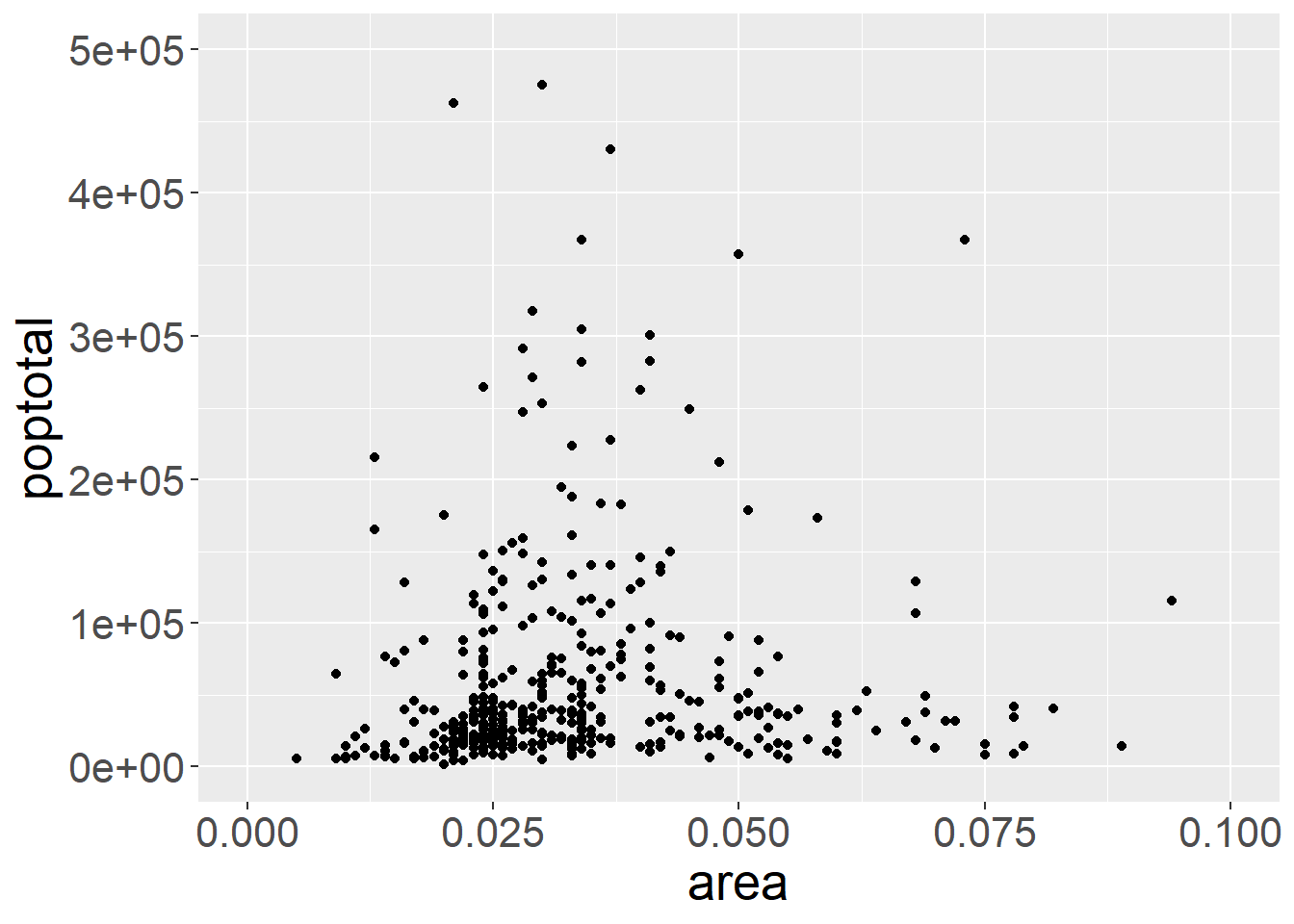
As mentioned before, providing extra arguments to aes tells ggplot to alter how each data point is visualised based on the value of that data point. Based on this we can also supply arguments like color and size:
ggplot(midwest, aes(x=area, y=poptotal)) +
geom_point(aes(color=state, size=popdensity)) +
xlim(c(0, 0.1)) + # plot inside a range that suit us
ylim(c(0, 500000)) +
theme(text = element_text(size=20)) #make axis bigger
## Warning: Removed 15 rows containing missing values or values outside the scale range
## (`geom_point()`).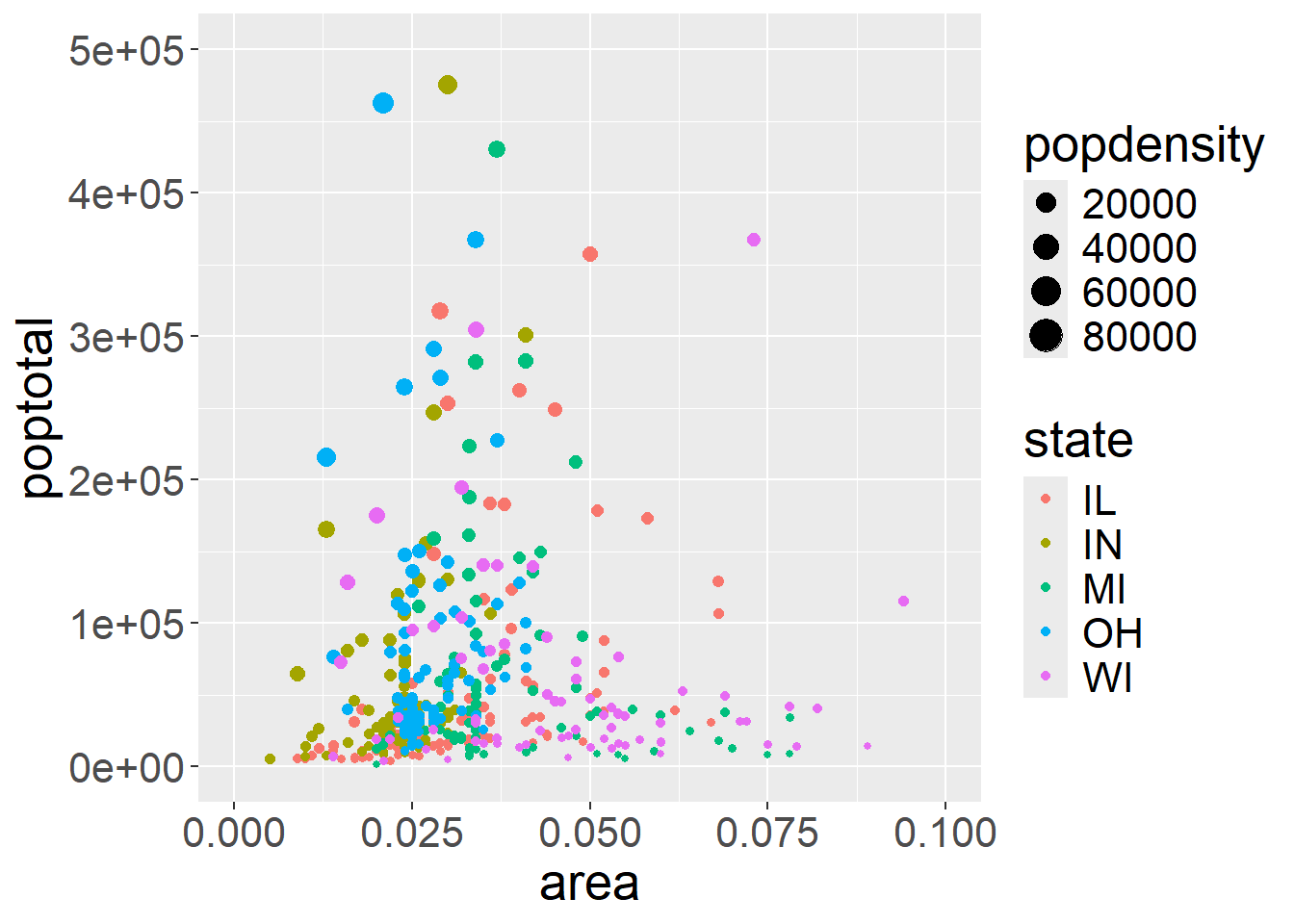
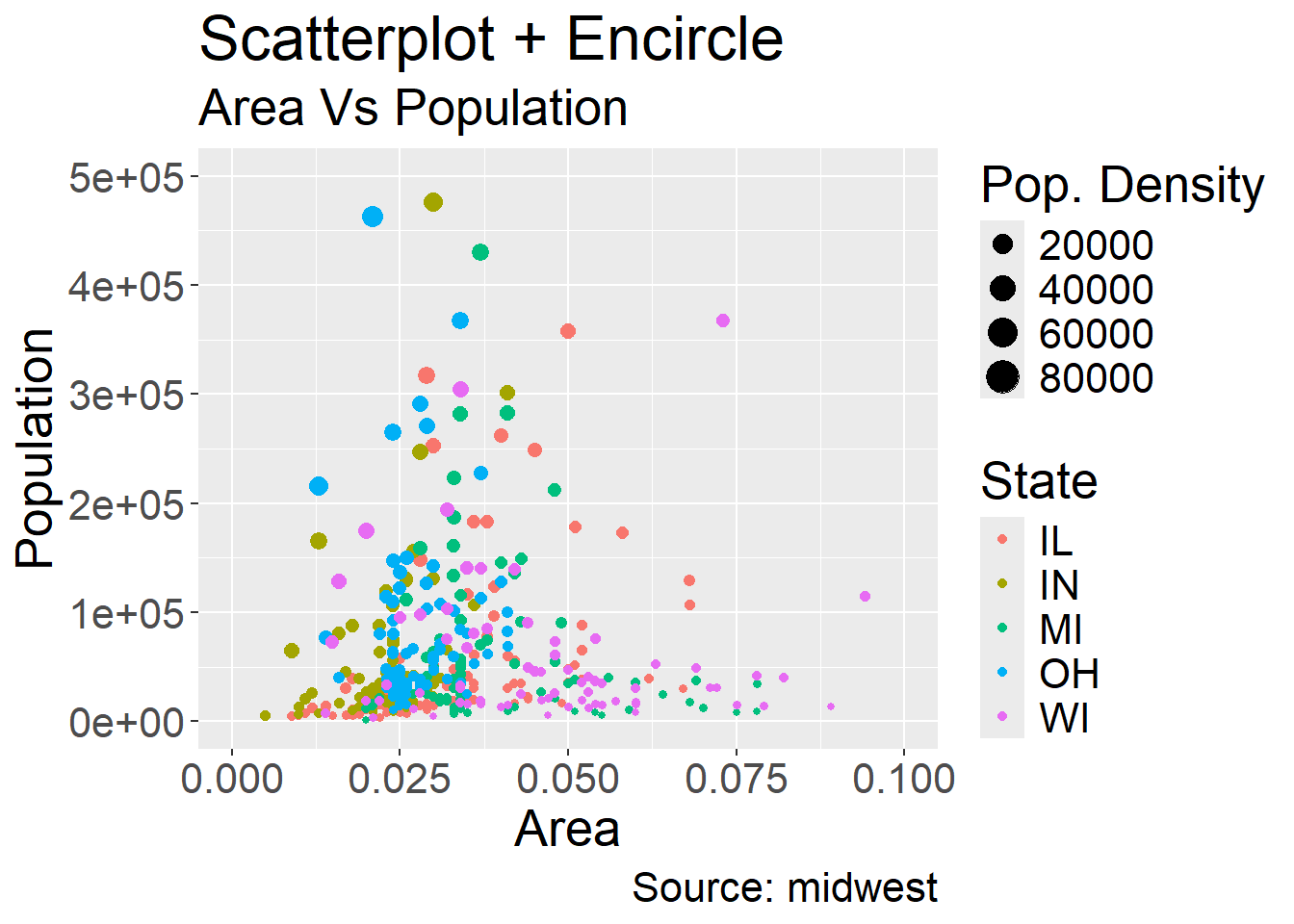
Conveniently, this also creates a legend for us by default however the naming is quite poor. We can fix this yet again using the labs argument:
ggplot(midwest, aes(x=area, y=poptotal)) +
geom_point(aes(col=state, size=popdensity)) + # draw points
xlim(c(0, 0.1)) +
ylim(c(0, 500000)) + # draw smoothing line
labs(subtitle="Area Vs Population",
y="Population",
x="Area",
title="Scatterplot + Encircle",
caption="Source: midwest",
color="State",
size="Pop. Density") +
theme(text = element_text(size=20)) #make axis/labels bigger
## Warning: Removed 15 rows containing missing values or values outside the scale range
## (`geom_point()`).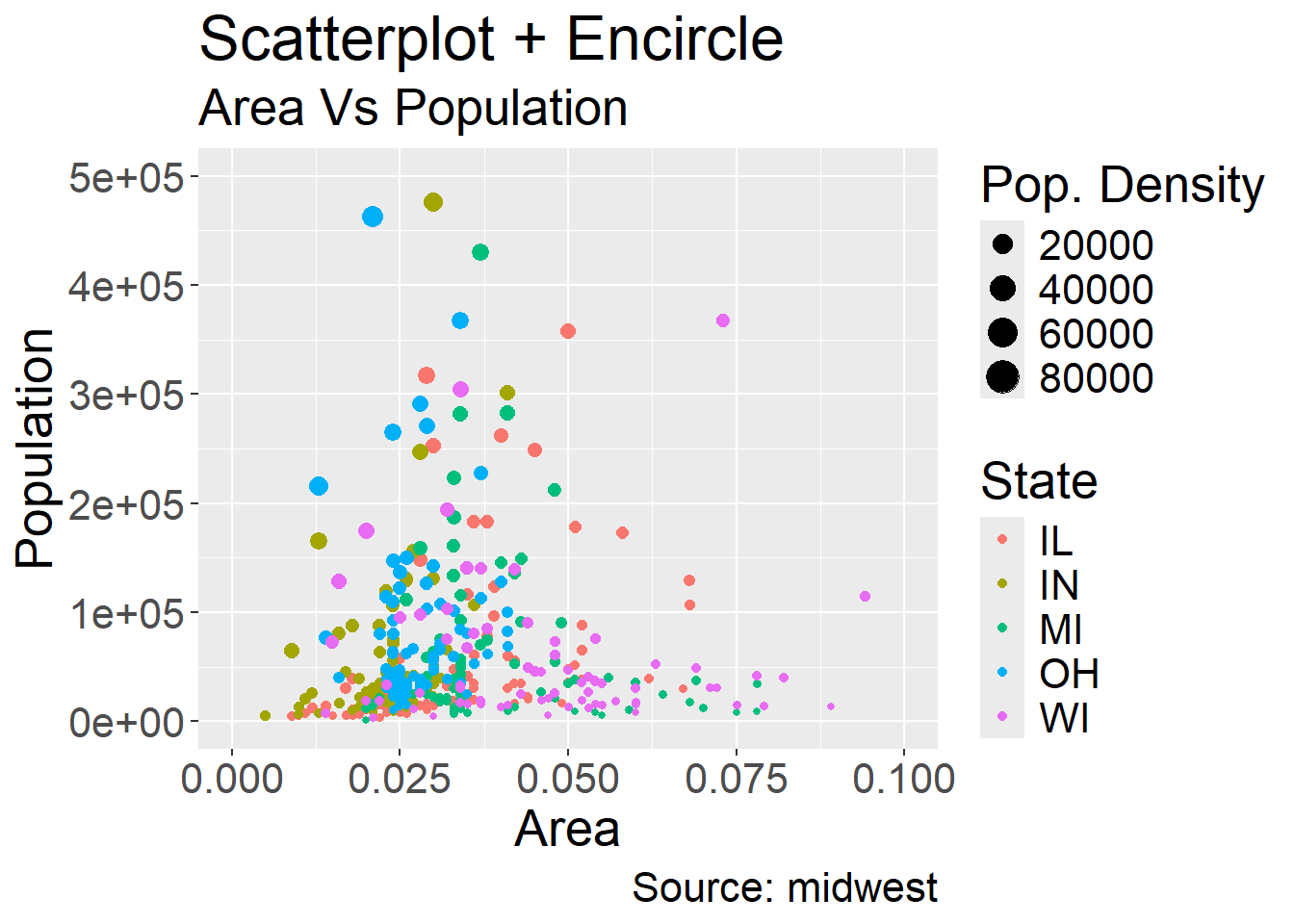
There are also other aesthetics that are often used for similar purposes:
alpha indicates numerical values as well by adjusting the transparencyshape indicates categorical values by adjusting the shape of the pointsfill works similarly to colour but for other geoms. For example, in a bar plot, fill is the colour of the bar, while colour is the border colourIn addition to a simple scatter plot, it may also be useful to know the number of observations at each point, and the marginal distributions:
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'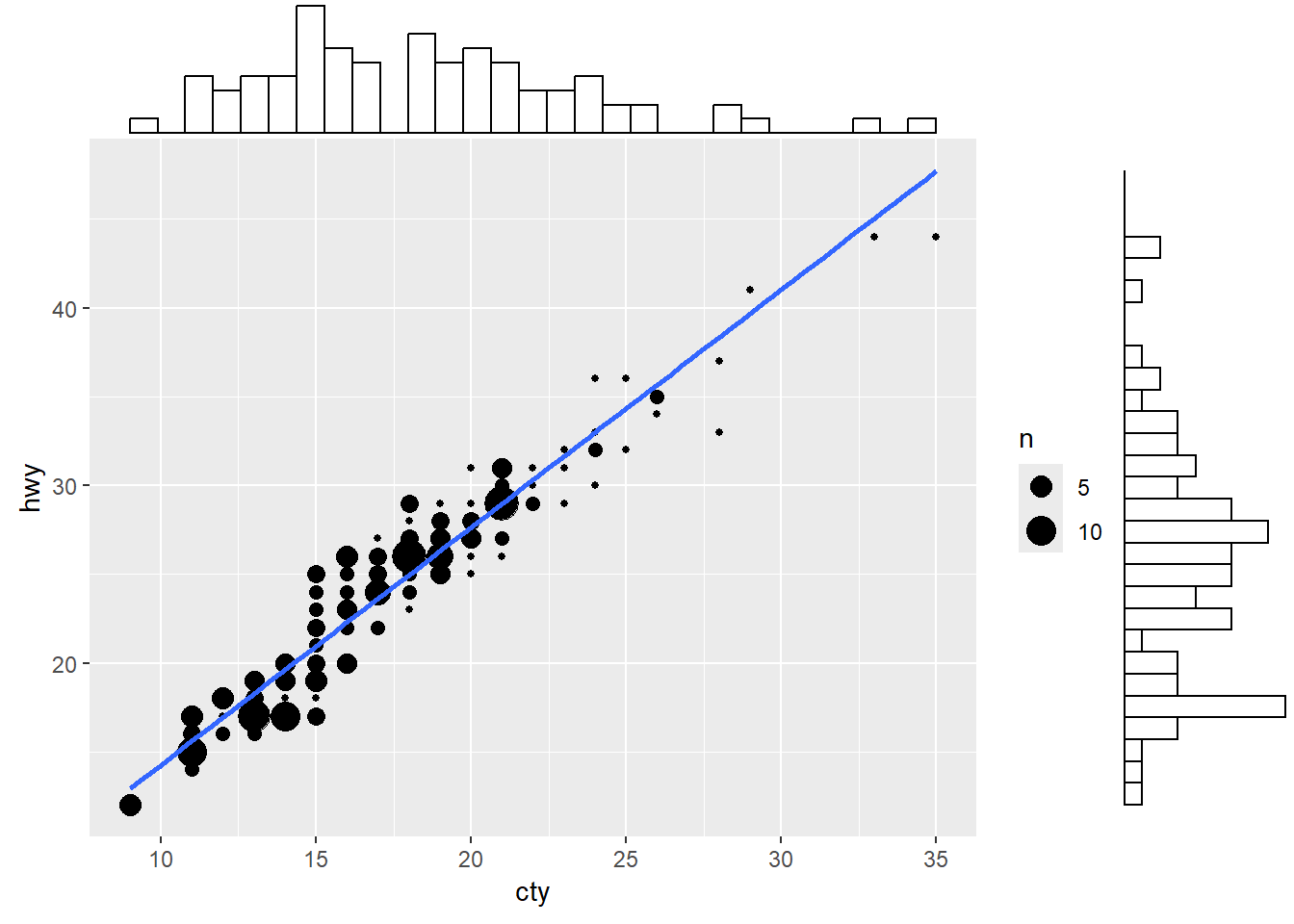
We start from a standard scatter plot of variable hwy vs cty (highway and city ‘miles per gallon’, for various car models) from data set mpg (included in the ggplot2 package).
ggplot(mpg, aes(cty, hwy)) +
geom_point()+
geom_smooth(method="lm", se=F) +
theme_bw()
## `geom_smooth()` using formula = 'y ~ x'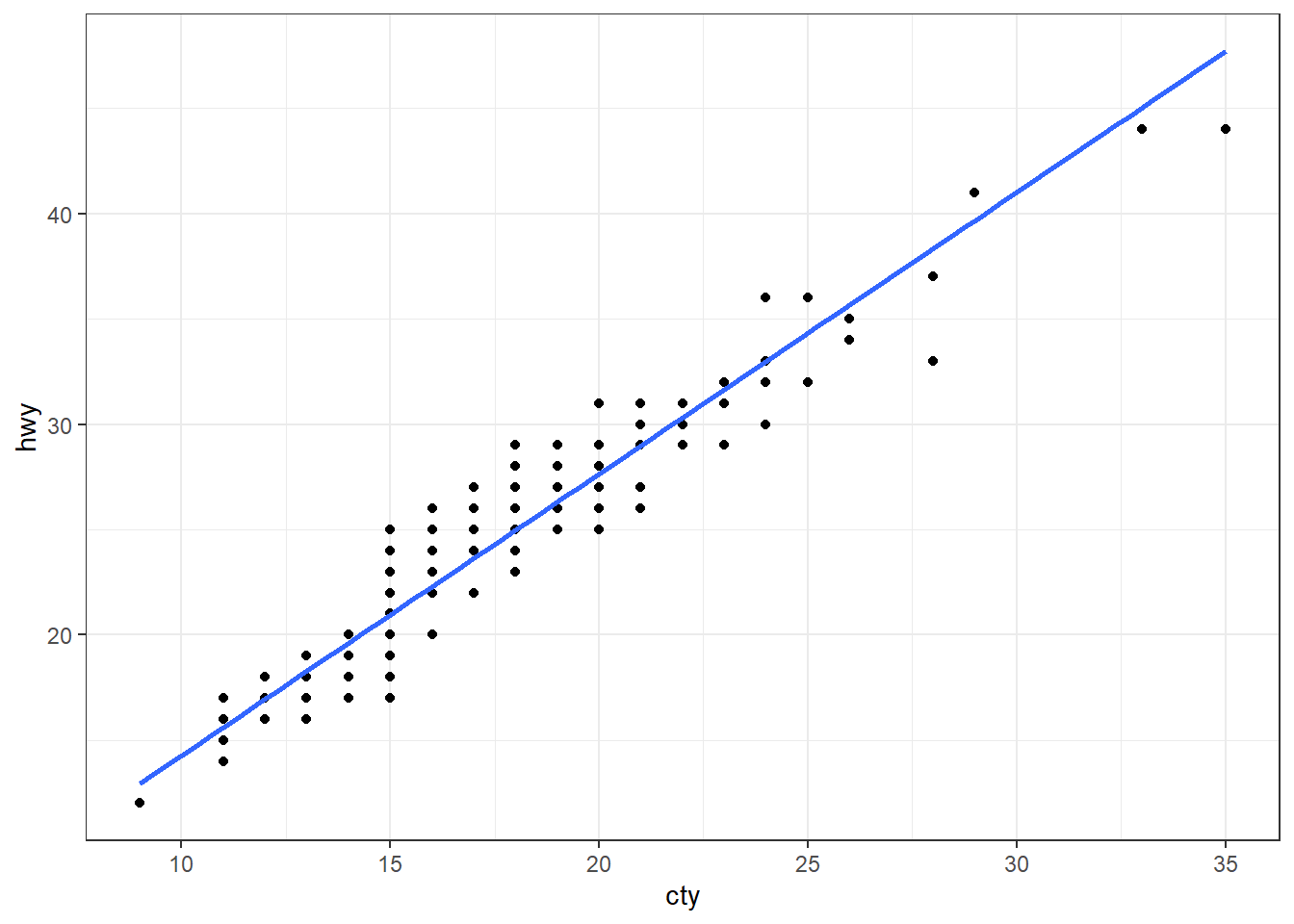
Have a look at the data set. Are cty and hwy truly continuous variables? What problem does this create?
head(mpg, 10)
## # A tibble: 10 × 11
## manufacturer model displ year cyl trans drv cty hwy fl class
## <chr> <chr> <dbl> <int> <int> <chr> <chr> <int> <int> <chr> <chr>
## 1 audi a4 1.8 1999 4 auto… f 18 29 p comp…
## 2 audi a4 1.8 1999 4 manu… f 21 29 p comp…
## 3 audi a4 2 2008 4 manu… f 20 31 p comp…
## 4 audi a4 2 2008 4 auto… f 21 30 p comp…
## 5 audi a4 2.8 1999 6 auto… f 16 26 p comp…
## 6 audi a4 2.8 1999 6 manu… f 18 26 p comp…
## 7 audi a4 3.1 2008 6 auto… f 18 27 p comp…
## 8 audi a4 quattro 1.8 1999 4 manu… 4 18 26 p comp…
## 9 audi a4 quattro 1.8 1999 4 auto… 4 16 25 p comp…
## 10 audi a4 quattro 2 2008 4 manu… 4 20 28 p comp…Many values of cty and hwy are repeated, hence many points overlap
This means the previous plot does not show all points
We can solve this by showing the number of observations at each spot, using the size of the dot as an indicator.
This is simply done by replacing geom_point() with geom_count().
ggplot(mpg, aes(cty, hwy)) +
geom_count() +
geom_smooth(method="lm", se=F) +
theme_bw()
## `geom_smooth()` using formula = 'y ~ x'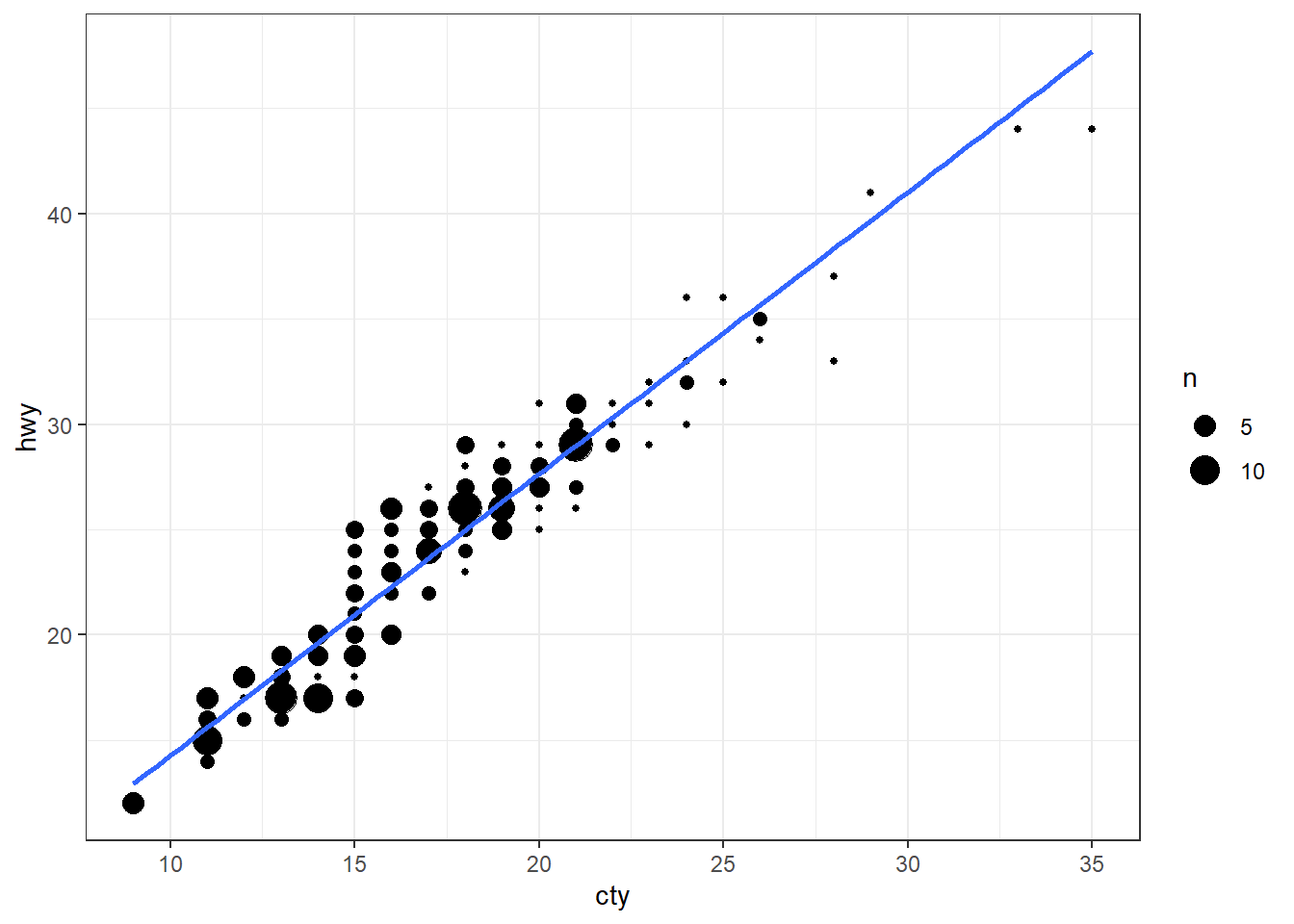
A scatter plot illustrates how two variables are related, but it could be useful to have a quick idea of their individual (marginal) distributions as well.
ggMarginal() function from the ggExtra package allows to visualise marginal distributions on top of a scatter plot. Below we add a histogram of marginal distributions on top of the scatter plot.g <- ggplot(mpg, aes(cty, hwy)) +
geom_count() +
geom_smooth(method="lm", se=F) +
theme_set(theme_bw())
ggMarginal(g, type = "histogram", fill="transparent")
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'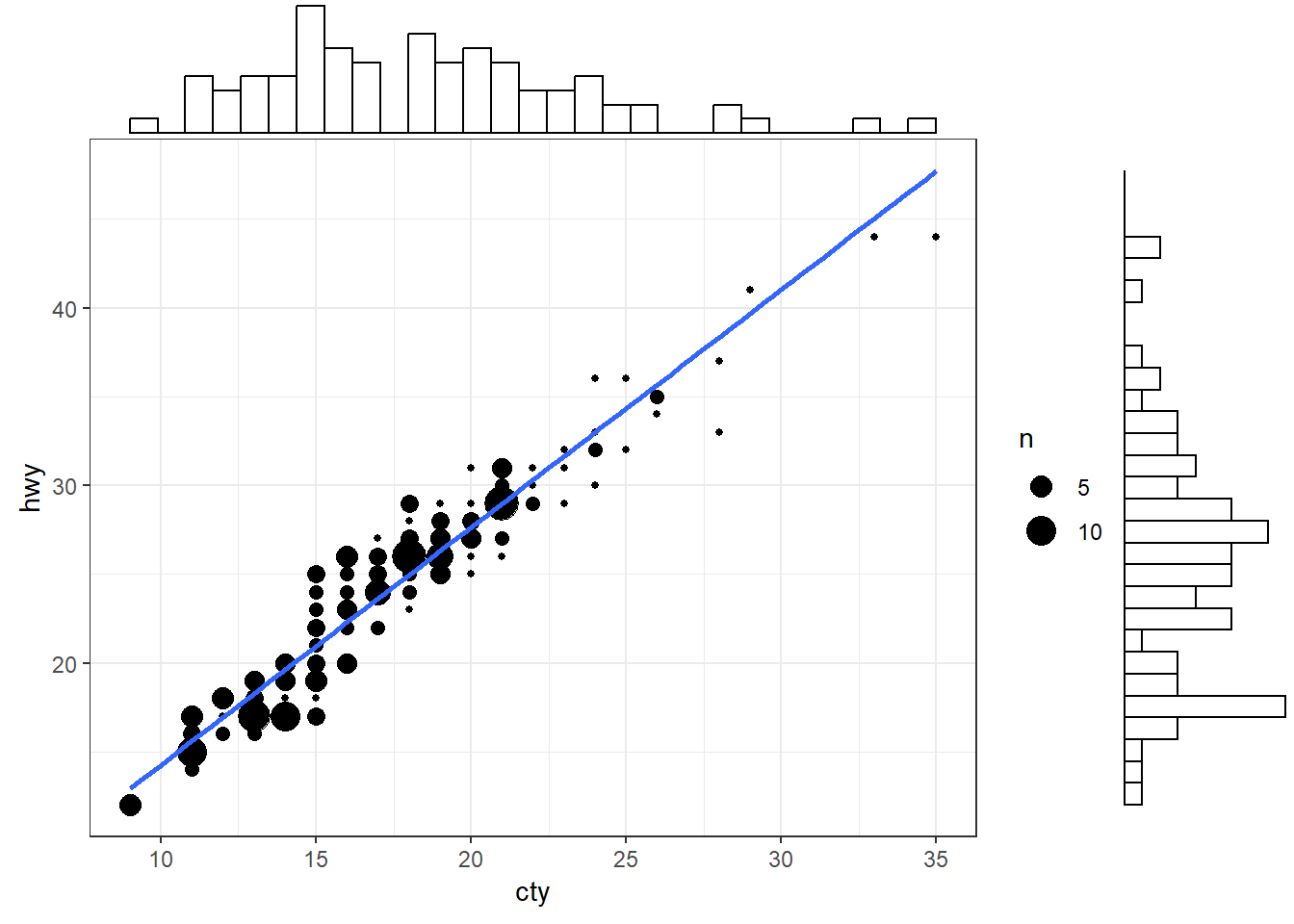
g <- ggplot(mpg, aes(cty, hwy)) +
geom_count() +
geom_smooth(method="lm", se=F)+
theme_set(theme_bw())
ggMarginal(g, type = "density", fill="transparent")
## `geom_smooth()` using formula = 'y ~ x'
## `geom_smooth()` using formula = 'y ~ x'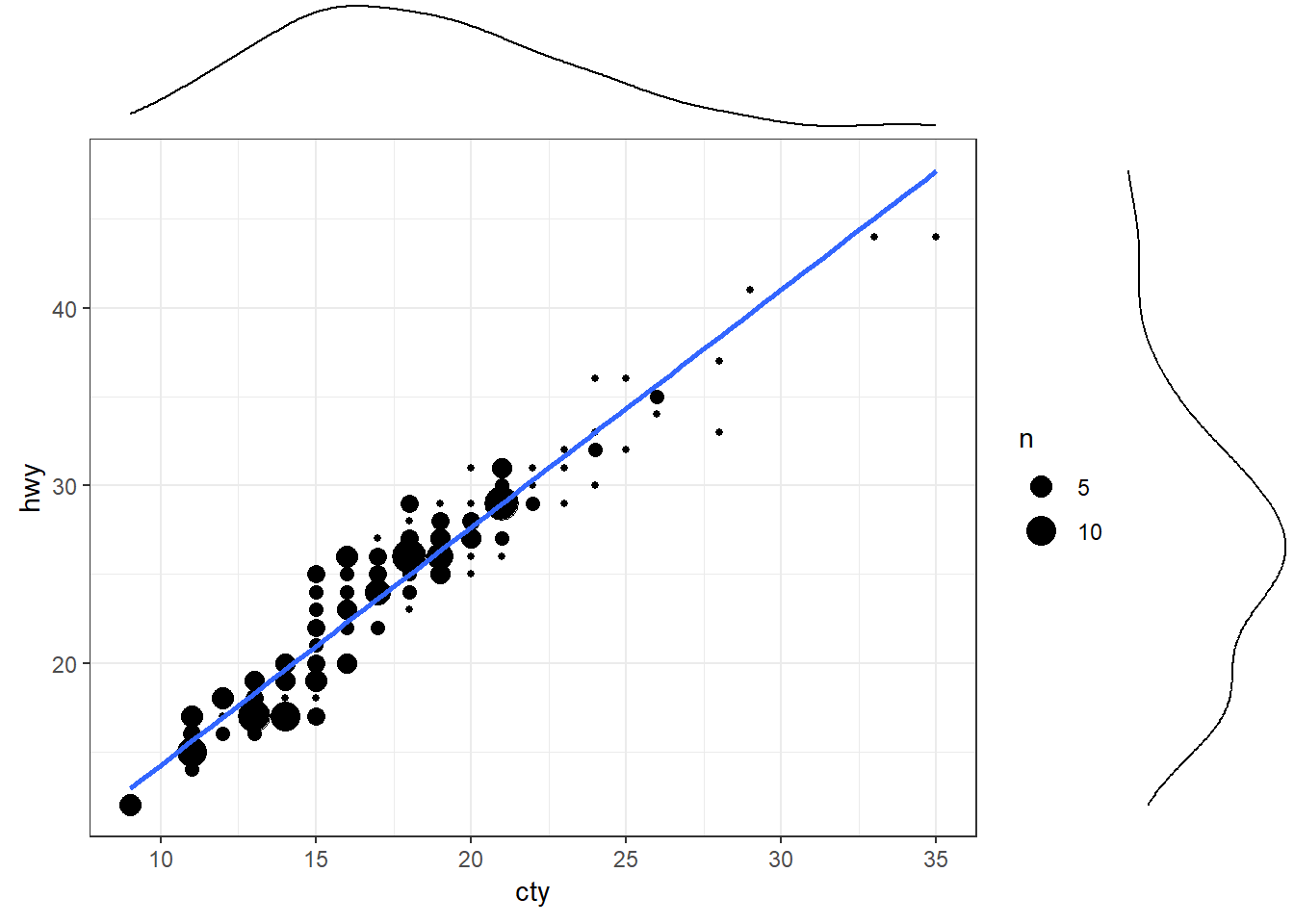
Coming back to the example we gave for “Other Aesthetics”, sometimes it is also useful to be able to encircle or highlight some data:
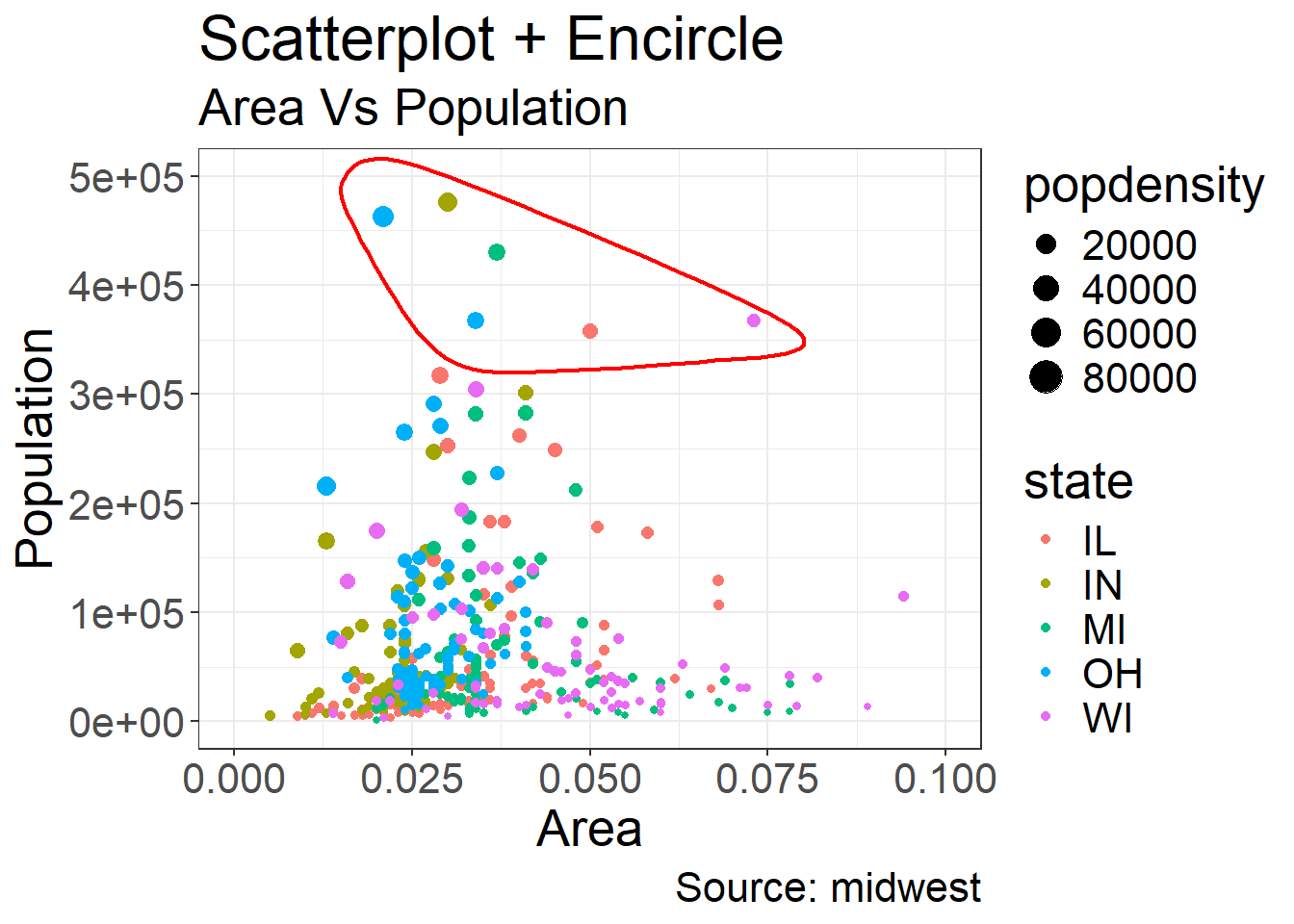
Coming back to the example we gave for “Other Aesthetics”, suppose that one is interested in emphasizing a certain area, then a range of dots can be circled
This is achieved by using geom_encircle in the ggalt package
Here we want to emphasize the observations where the population total is between 350k and 500k and where the area is between 0.01 and 0.1
Note that we need to find out these observations, which is done by creating the subset observations of midwest_select
We have also added the titles and caption
midwest_select <- midwest[midwest$poptotal > 350000 &
midwest$poptotal <= 500000 &
midwest$area > 0.01 &
midwest$area < 0.1, ]
ggplot(midwest, aes(x=area, y=poptotal)) +
geom_point(aes(col=state, size=popdensity)) + # draw points
xlim(c(0, 0.1)) +
ylim(c(0, 500000)) + # draw smoothing line
geom_encircle(data=midwest_select,
color="red",
size=2,
expand=0.08) + # encircle
labs(subtitle="Area Vs Population",
y="Population",
x="Area",
title="Scatterplot + Encircle",
caption="Source: midwest") +
theme(text = element_text(size=20)) #make axis/labels bigger
## Warning: Removed 15 rows containing missing values or values outside the scale range
## (`geom_point()`).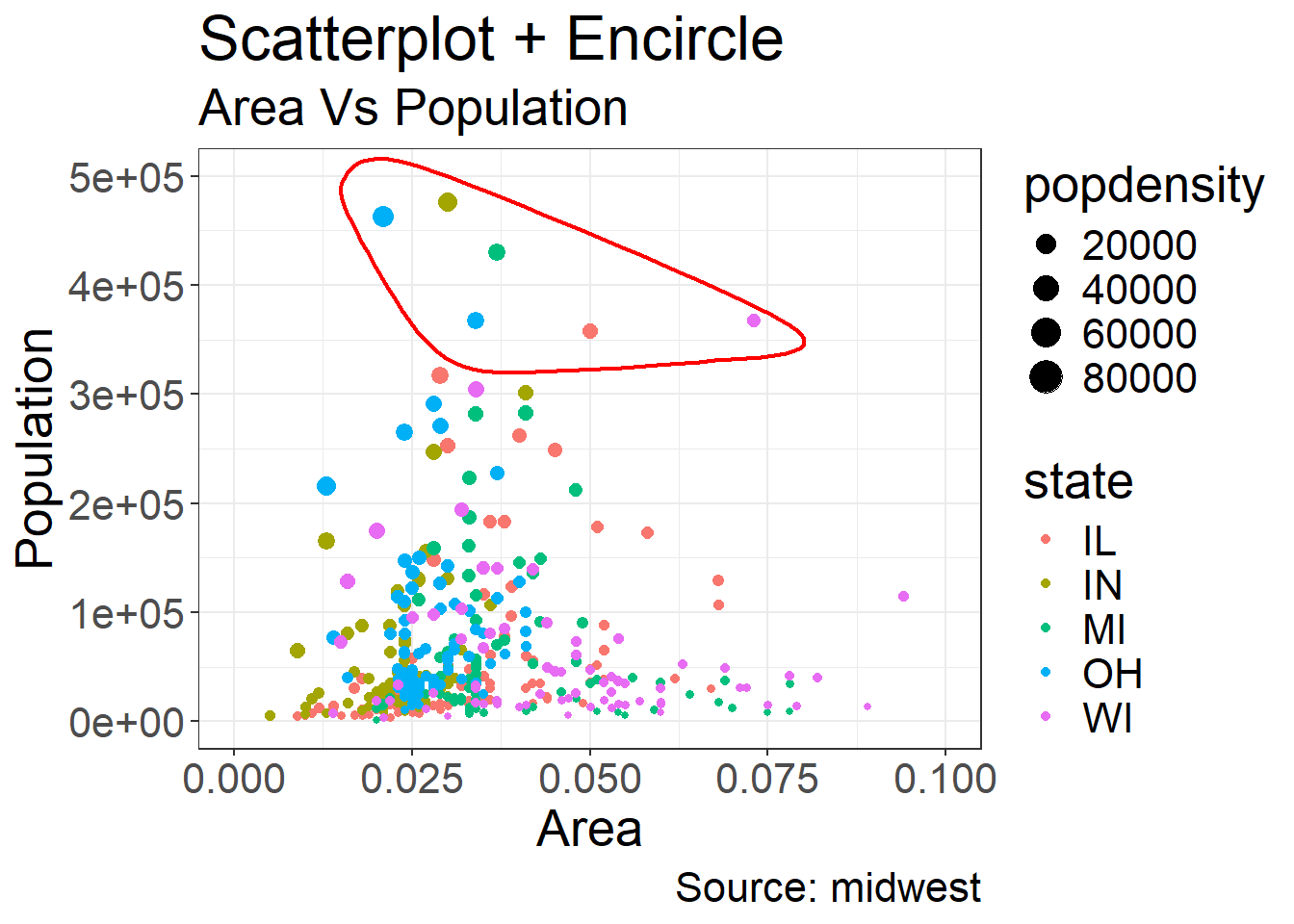
ggplot can also be used to create thematic maps to visualise data based on geography.
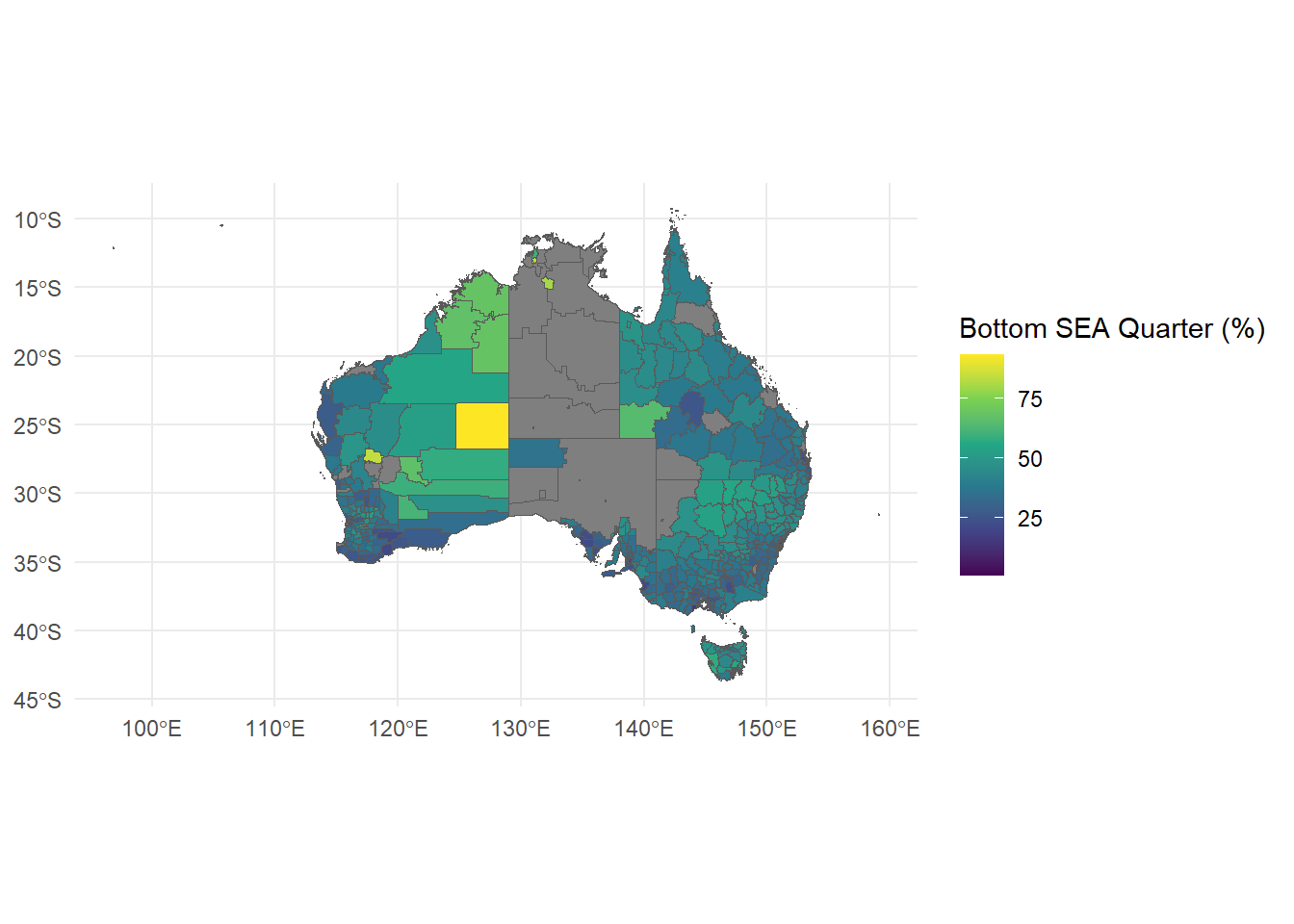
The first step is to import map data. These kinds of datasets are called “shapefiles” because they are datasets that help programs like R draw shapes by spelling out complex polygons as a list of points. In this case the map is of Australia broken down into its Local Government Areas (LGA).
LGA11aAust.shpLGA11aAust.cpgLGA11aAust.dbfLGA11aAust.shxLGA11aAust.prjOnce geometry data is found, we can use geom_sf (sf for shapefile) to create a map image:
To make this graph informative, we then need some numerical data which is also grouped by LGA to plot on this map. heatmap_data.csv contains data on Index of Community Socio-Educational Advantage (ICSEA) by each LGA, so we need to merge this data with our shapefile data.
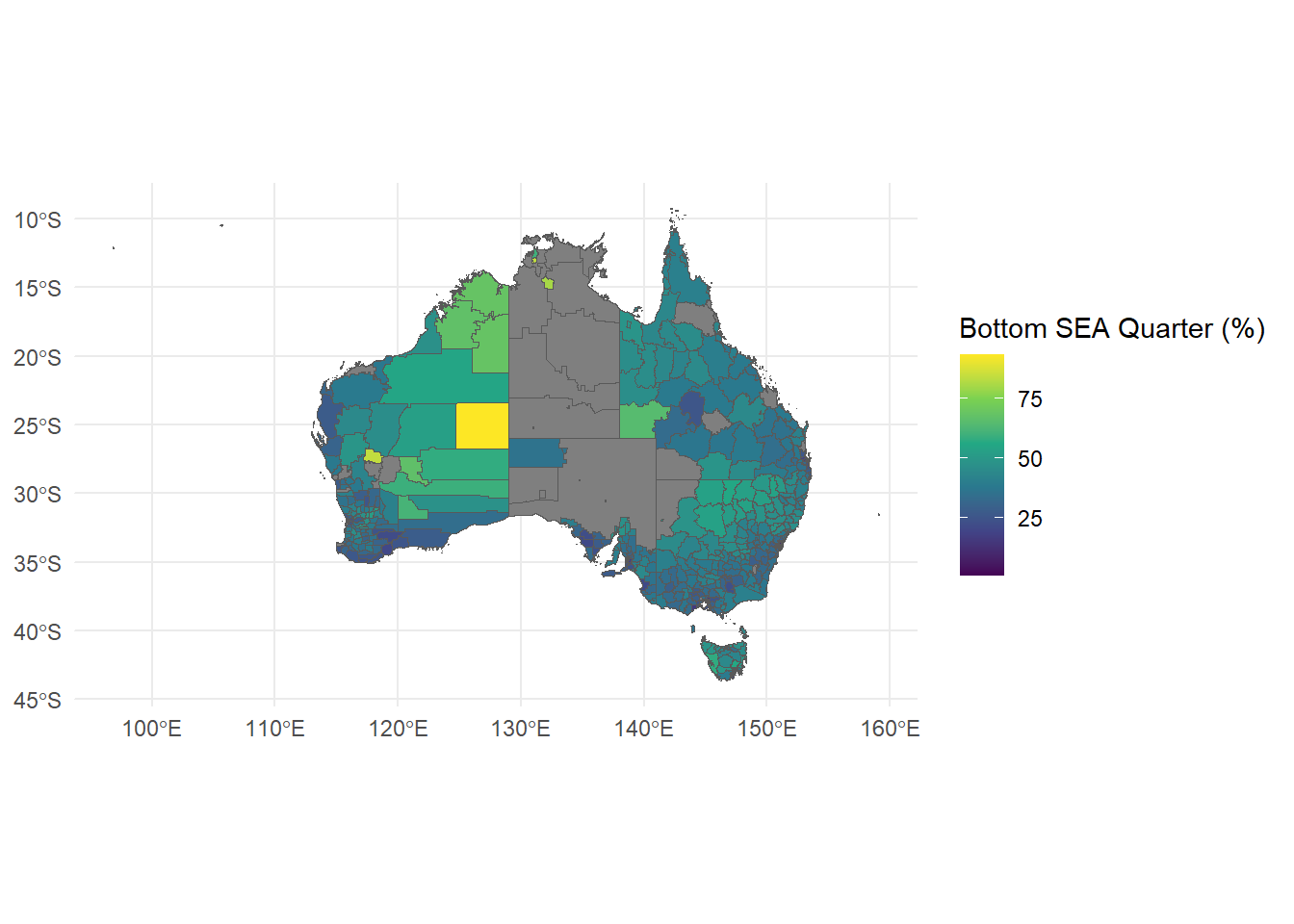
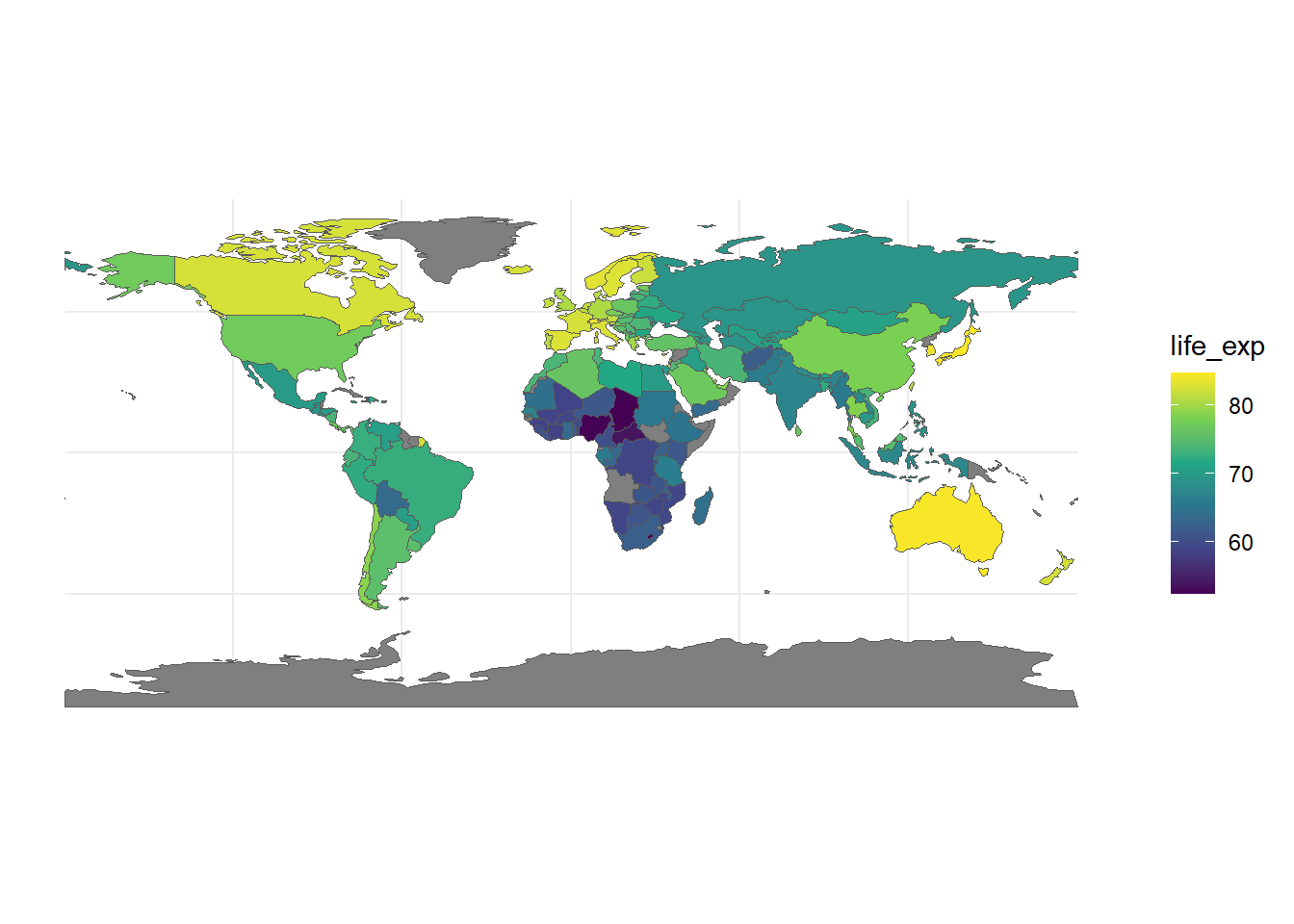
tmap which is more specifically designed for this task and comes with some nice default features.tidyverse.Use the world.gpkg data set to generate a map of the world using life expectancy as the scale. .gpkg files are another way of storing shapefile data other than the 5 file method you saw before but you can read it the same way using st_read(world.gpkg).
You can either download the dataset from Ed or by clicking here.
This data set is fully created by the publishers of tmap.
Use the last_plot() to returns the last plot you have created.
One can use the ggsave() function to save the last plot you have created.
One can use the function qplot() in a similar way as a traditional R plot function (but without the ‘plus sign’ synthax).
qplot(height,weight, data = raw_data)
## Warning: `qplot()` was deprecated in ggplot2 3.4.0.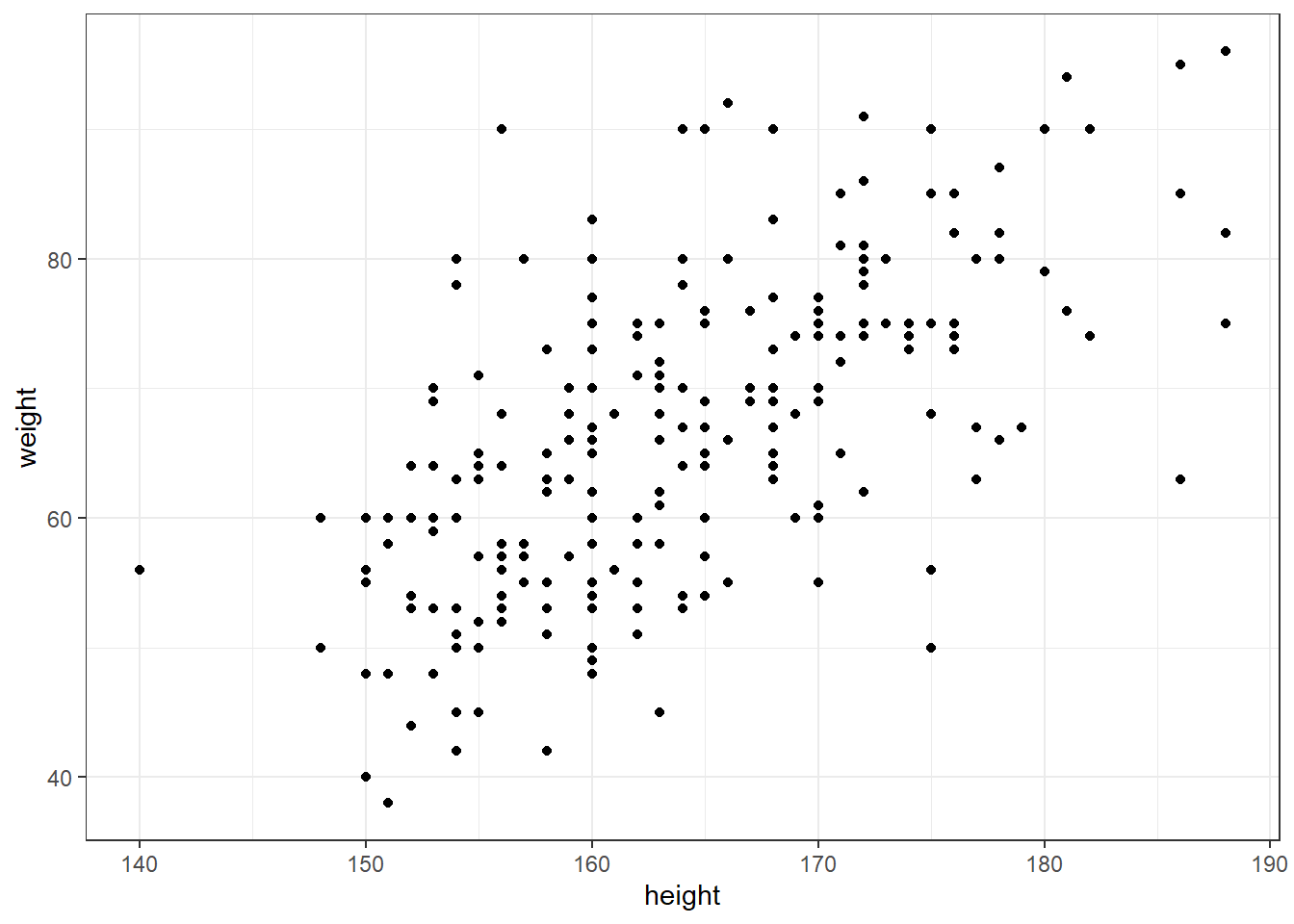
10 weeks unfortunately is simply not enough time to teach you all the amazing things that can be done with R.
With that in mind, the next slides will give you some ideas of various packages and frameworks you can look into if you are interested. These are all optional and just for those that are interested.
Obviously not all of these will interest all of you, but if there is one that you are intrigued by, then it is always a good idea to investigate further.
These are not in any particular order, some are very large and complex tools, and some are packages for some specific but useful purposes.
purrr and tidyr are two packages that are part of the tidyverse that can make your code much more efficient when used well. The first allows you to use functional programming within R (if you are unsure of what this is the website explains it a bit) and the second is for tidying and reshaping data.tidyquant is a package that aims to take all of the previously popular financial analysis packages in R and implement them in a tidyverse way. This allows for greater integration of financial analysis with other tools like ggplot2.plotly. While ggplot2 is excellent for static plots and has huge customisability, it tends to be difficult to create more “flashy” graphs like 3D plots or interactive plots. plotly aims to go the other way by having plots that are interactive, allowing for 3D plots, and by default including some “fancy” types of plots. You should go check out some examples on their site.ggraph is an extension of ggplot2 which provides an easy way to create plots of relational data structures like networks, graphs and trees. This is definitely more of a specialty use case, but if you do need to draw graphs of graphs, this is an incredibly helpful package.tidymodels is a package for modelling and machine learning, aiming to follow the tidyverse principles as well. This package may not be very useful yet as you would not have learned much about modelling (you have to wait for a course like ACTL3142 for that). However, once you are doing modelling, tidymodels is extremely well-designed and prevents the need for using a ridiculous number of packages all to do slightly different things.data.table is a tool for manipulating large datasets. It performs a similar purpose to dplyr but with a couple of key trade-offs. data.table tends to have more confusing and unintuitive syntax and is not well integrated with the rest of the tidyverse. However, it is significantly faster than dplyr, particularly on large datasets, and is very memory efficient. If you are working with large datasets, it is worth looking into.R can work with large datasets, at a certain point it becomes difficult and databases are significantly more efficient.dplyr in how it functions (select, filter, group by, etc.).R feel free to look into SQLite, but for the majority that are not as comfortable and want to learn how to use databases within R, the R4DS textbook has a great introduction to databases through R, linked here.RMarkdown, which is a tool that allows you to write documents using parts of the \(\LaTeX\) engine, but more importantly allows you to embed R code that can be run into your documents. This allows you to create documents that can be easily updated with new data or new code, and the results will automatically update in the document. Demonstrating its abilities is best done through examples, so please check out the gallery and if you are interested in learning more this page has many places for you to get started (as always, it is R4DS to the rescue :)).RMarkdown is a great tool for writing documents, it is not the only one. Quarto is a multi-language, next generation version of R Markdown from the same people who made RStudio, R Markdown and Shiny. Quarto is much more flexible and powerful than R Markdown. In fact, the entire course website and slides for this course are all made with Quarto! Surely that must give you some motivation to learn it! Quarto effectively combines many popular tools for document/website/presentation creation and allows you to use them all in one. This means you can create websites using Shiny or ReactJS, presentations using Beamer or RevealJS, articles/books using Typst or \(\LaTeX\), dashboards and many more. As usual, examples show it best, and for getting started yet again it is R4DS that will serve you best. Once you understand the basics, the Quarto website has many guides to get you started with more complex use cases.A Shiny app is used to create web applications that incorporate interactive visualisations. It is hard to explain the full capabilities of Shiny, so you are better off looking at some examples.
You can find a good introduction to Shiny app on the official website of R Studio:
Here is a course on Shiny, by one of the authors of your R book (Pierre Lafaye de Micheaux):
Use the mtcars dataset in R and present the histogram and empirical distribution of the variable mpg.
# the number of bins needs to be relatively small here
# given the small size of sample
# log-transformation is not necessary here
ggplot(mtcars, aes(mpg))+geom_histogram(bins=5) # histogram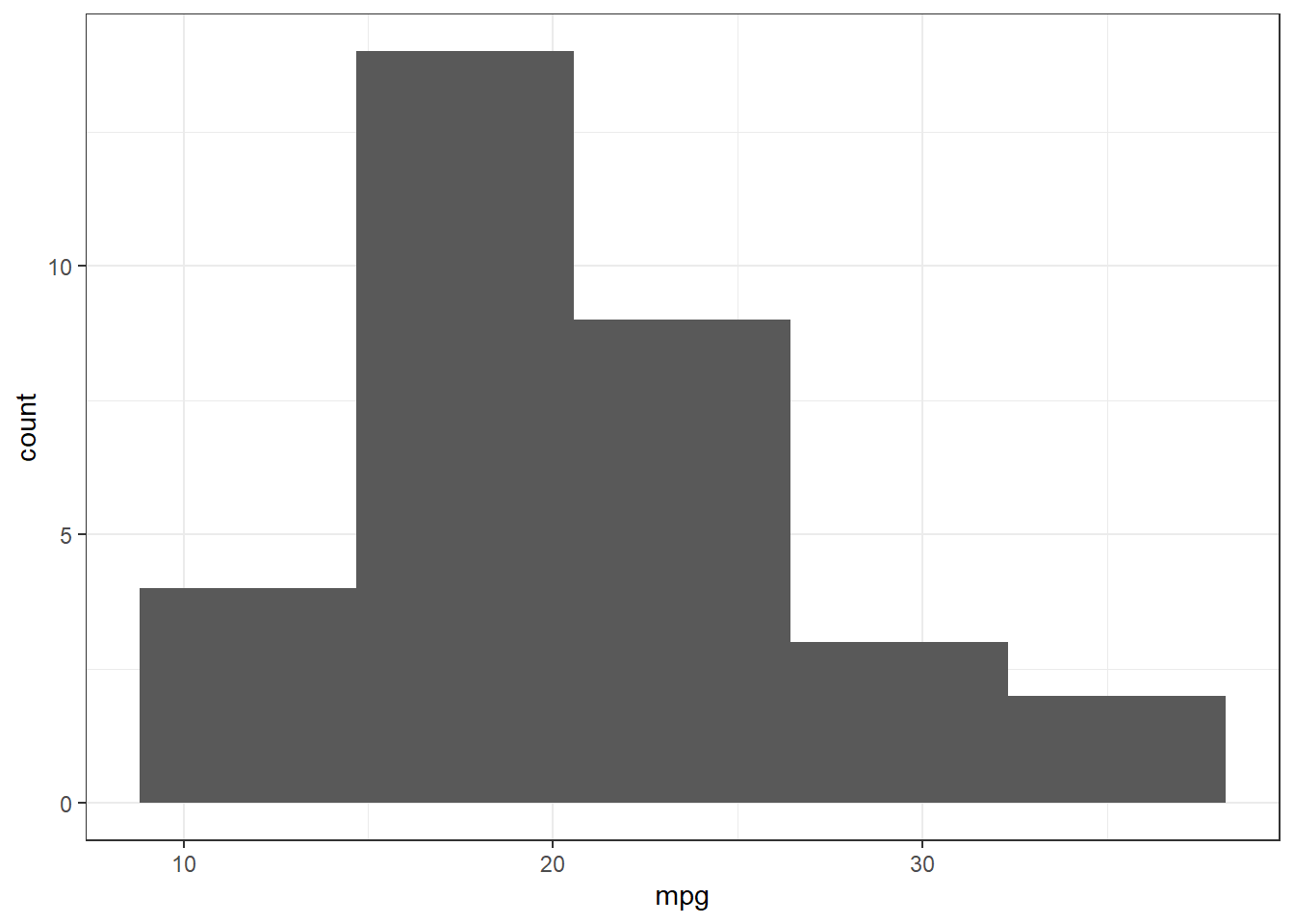
ggplot(mtcars, aes(mpg))+stat_ecdf() # empirical distribution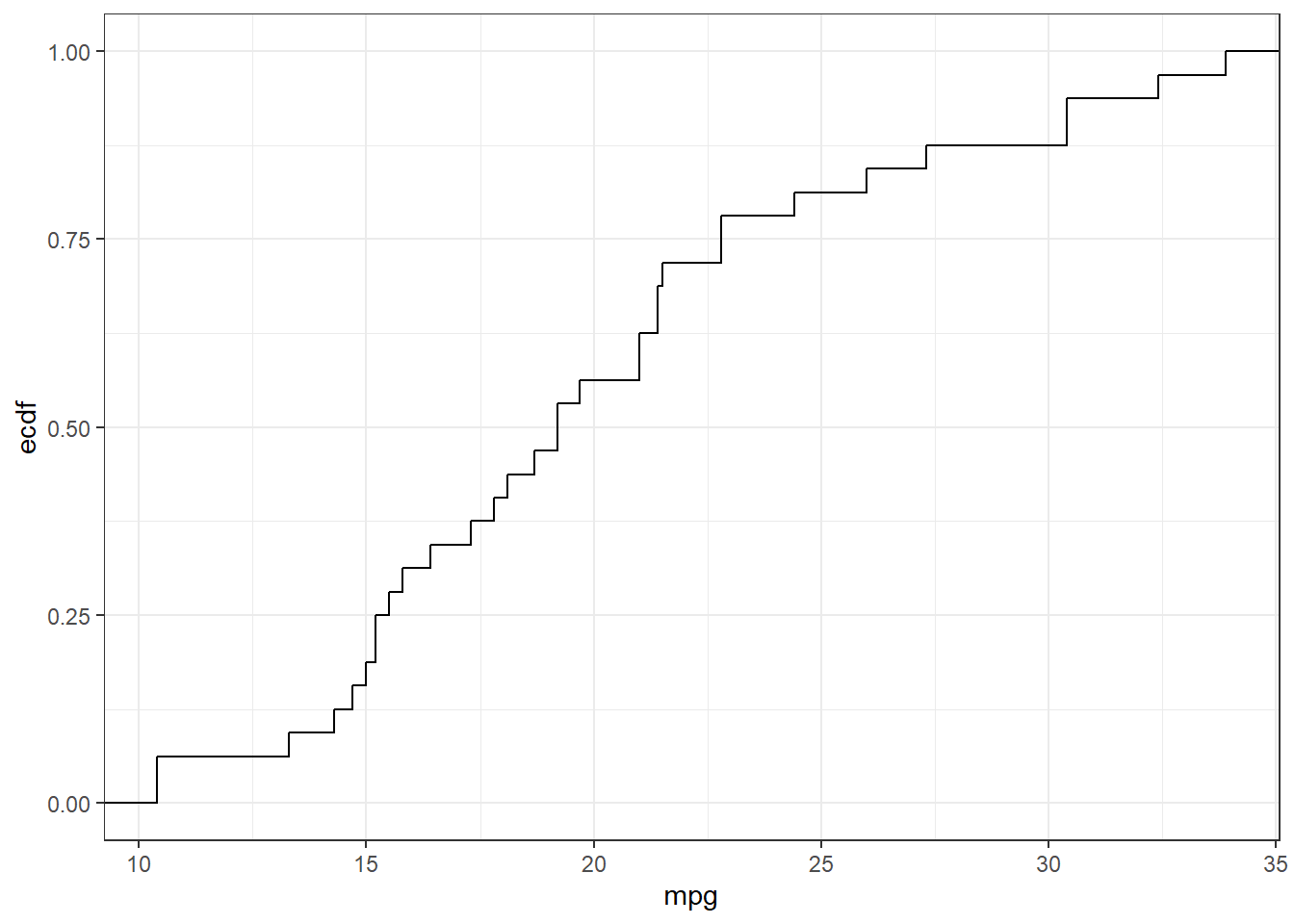
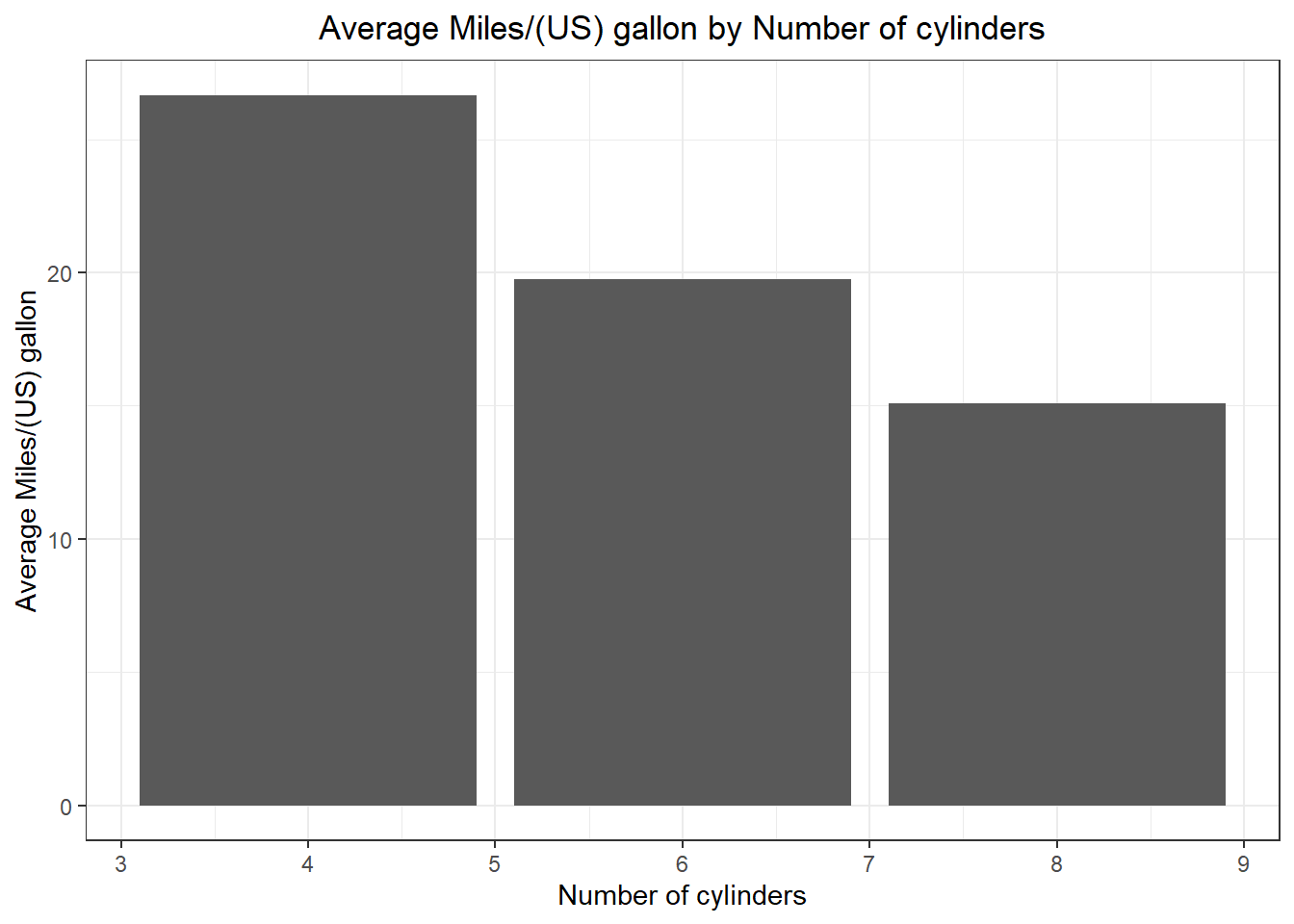
Use the mtcars dataset in R and present the average Miles/(US) gallon for each Number of cylinders.
# aggregate mpg given cyl
# the function we are interested in mean
mpg_cyl <- aggregate(mpg~cyl, mtcars, mean)
# create the bar plot
ggplot(mpg_cyl, aes(cyl, mpg))+
geom_bar(stat = "identity") +
labs(x = "Number of cylinders", y = "Average Miles/(US) gallon") +
ggtitle("Average Miles/(US) gallon by Number of cylinders")+
theme(plot.title = element_text(hjust = 0.5))