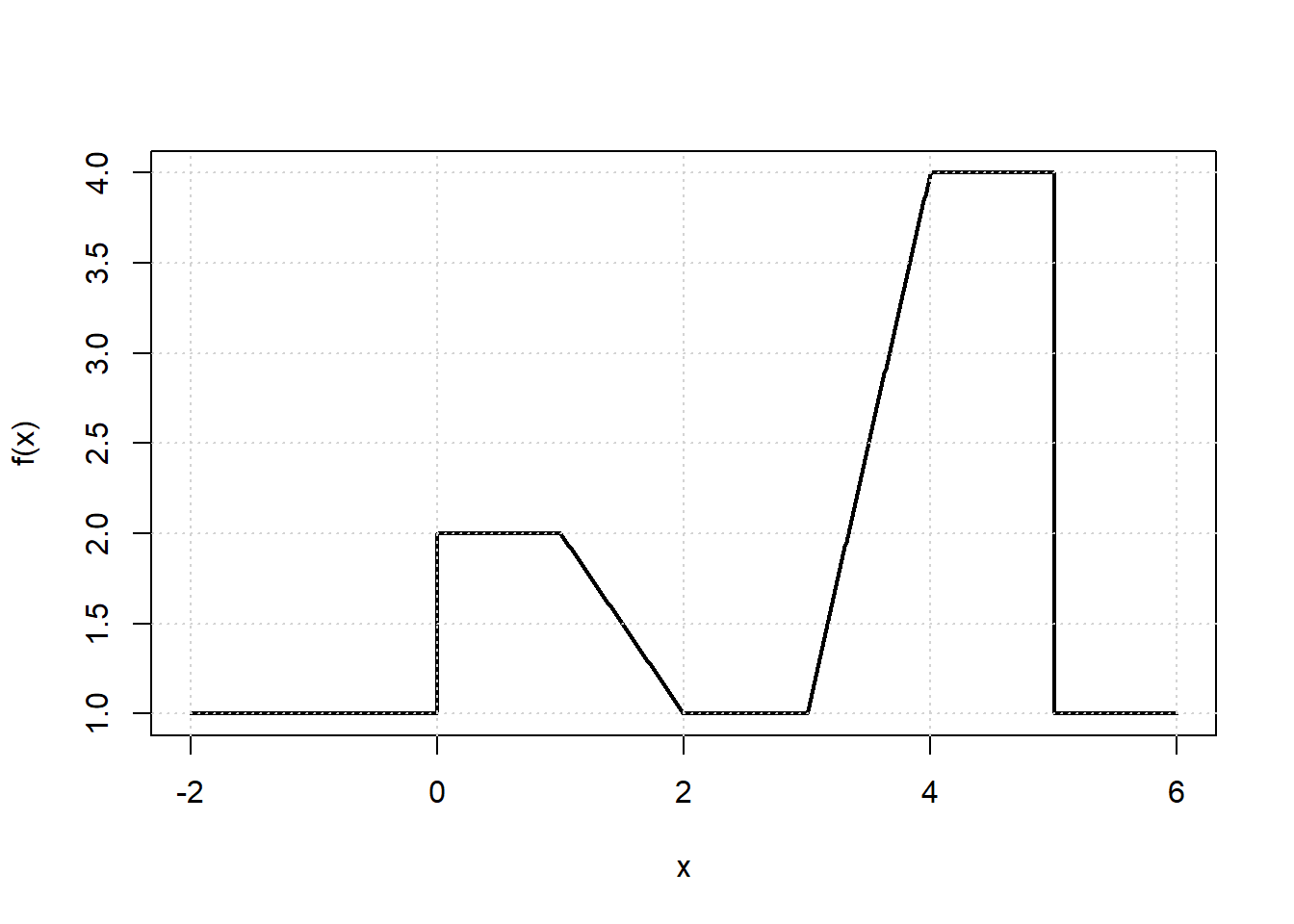
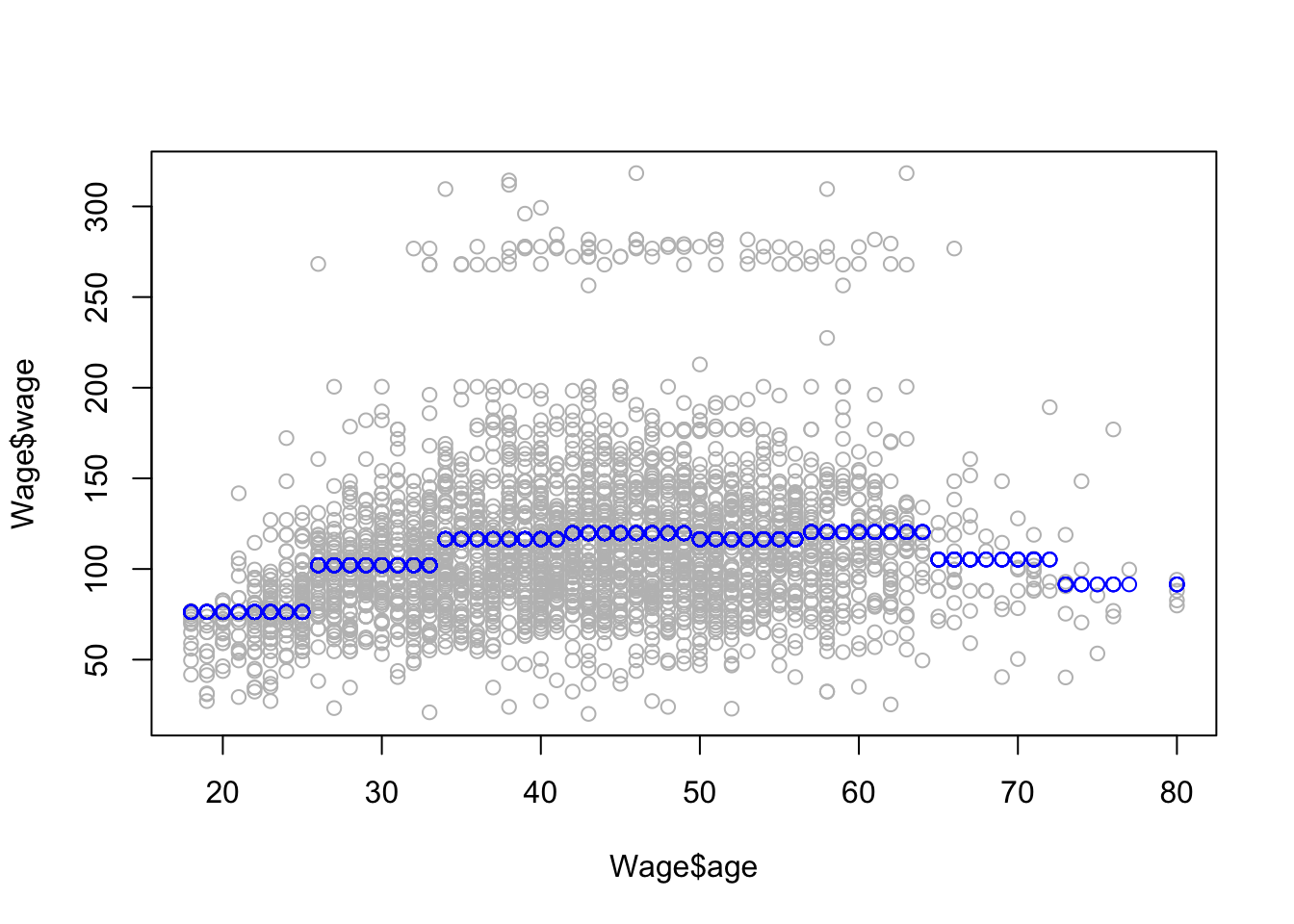
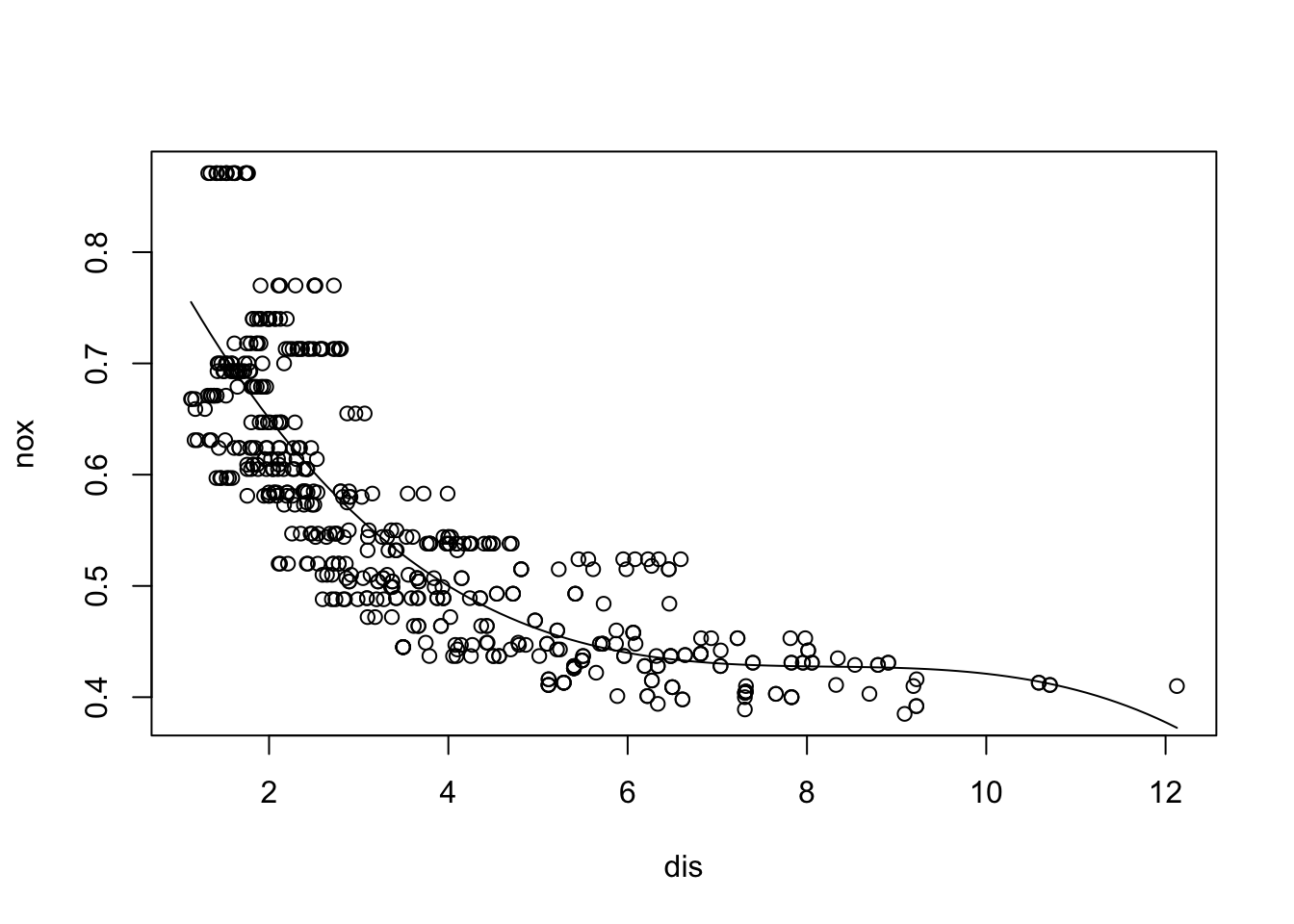
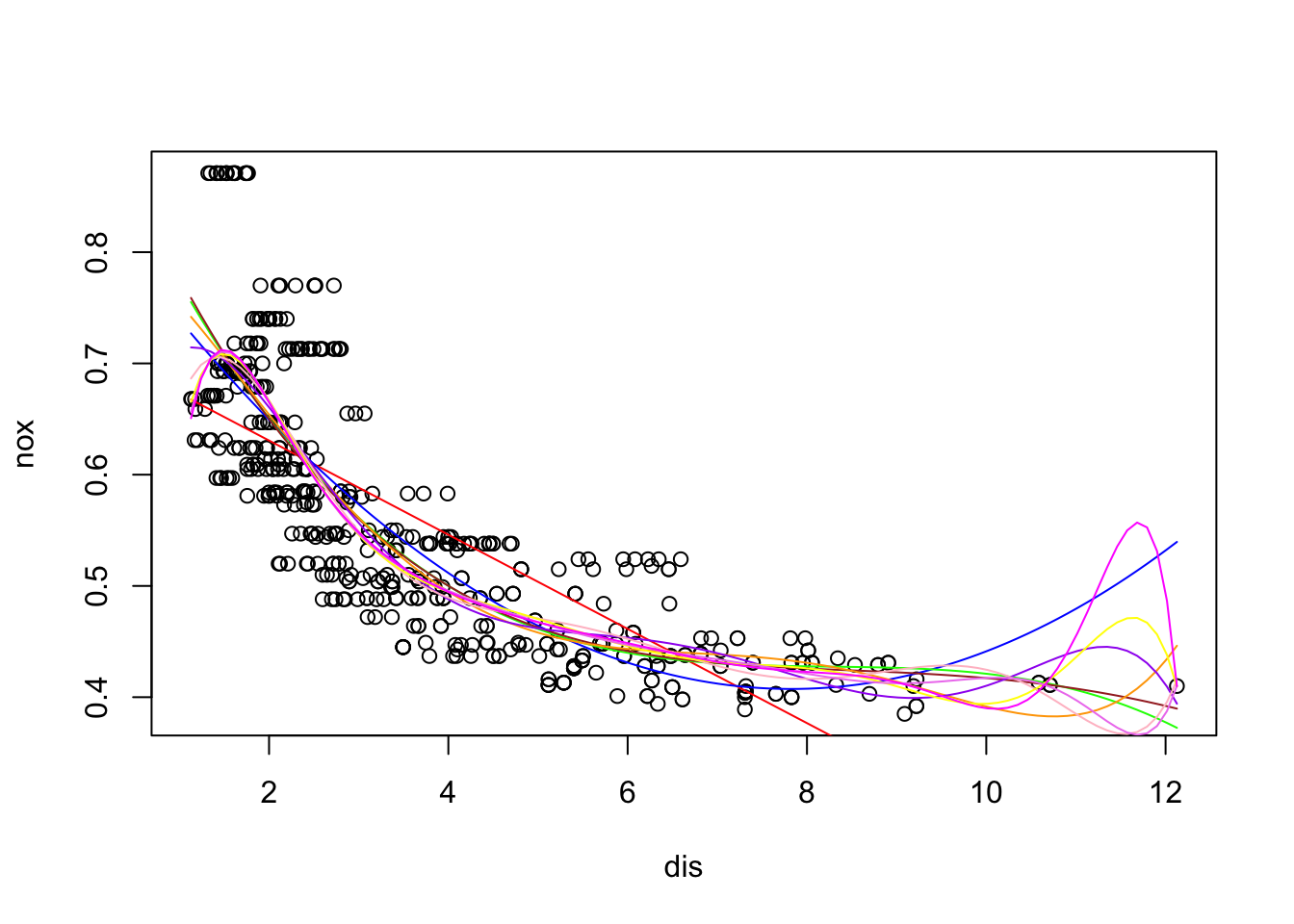
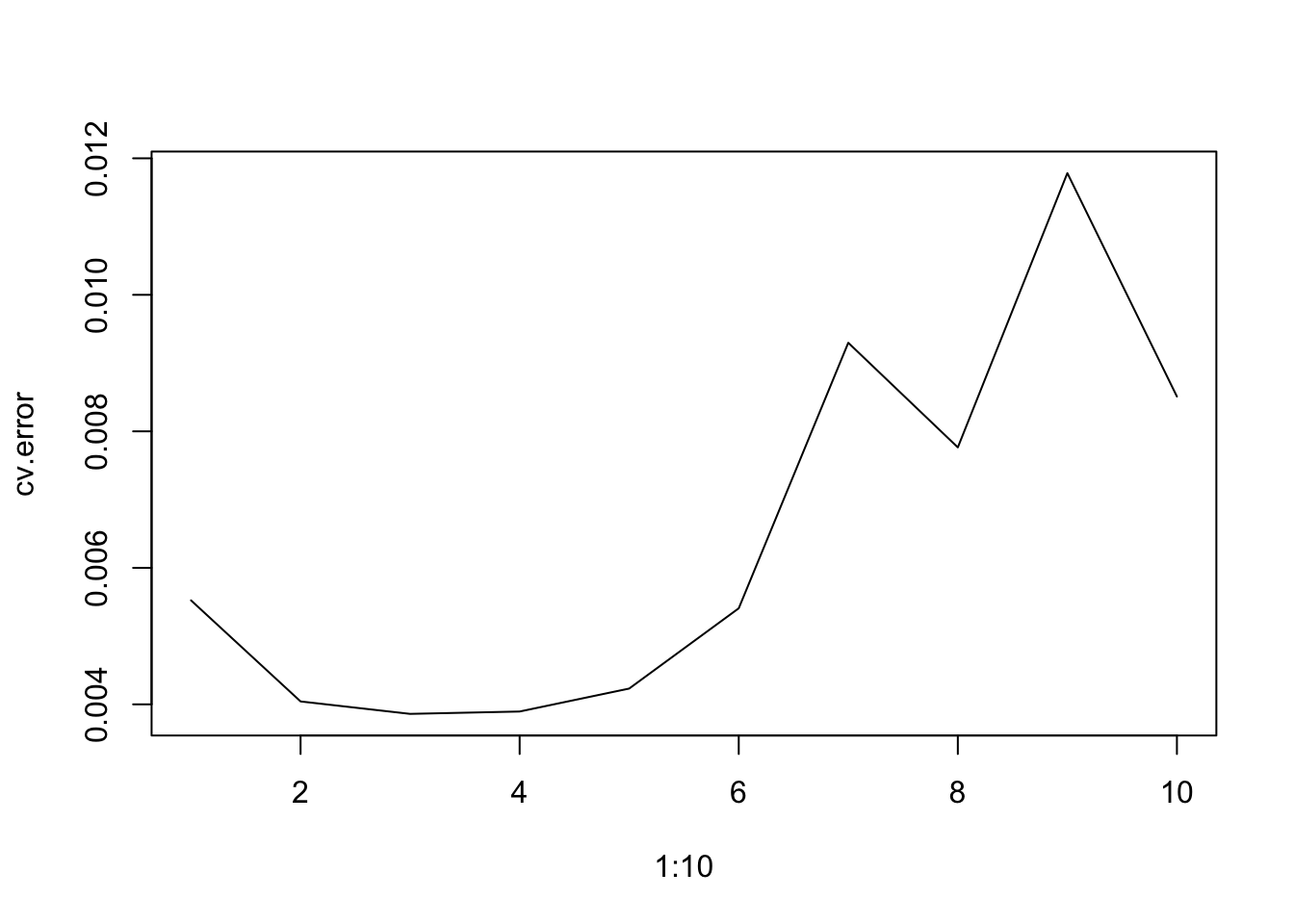
x <- seq(-2, 6, length.out = 1000)
b1 <- function(x) I(0 <= x & x <= 2) - (x - 1) * I(1 <= x & x <= 2)
b2 <- function(x) (x - 3) * I(3 <= x & x <= 4) + I(4 < x & x <= 5)
f <- function(x) 1 + 1 * b1(x) + 3 * b2(x)
plot(x, f(x), type = "l", lwd=2)
grid()