Show the R package imports
library(tidyverse)
library(splines)
library(mgcv)library(tidyverse)
library(splines)
library(mgcv)import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from patsy import dmatrix
import statsmodels.api as sm
import statsmodels.formula.api as smfSome of the figures in this presentation are taken from “An Introduction to Statistical Learning, with applications in R” (Springer, 2021) with permission from the authors: G. James, D. Witten, T. Hastie and R. Tibshirani.
James et al. (2021): Chapter 7
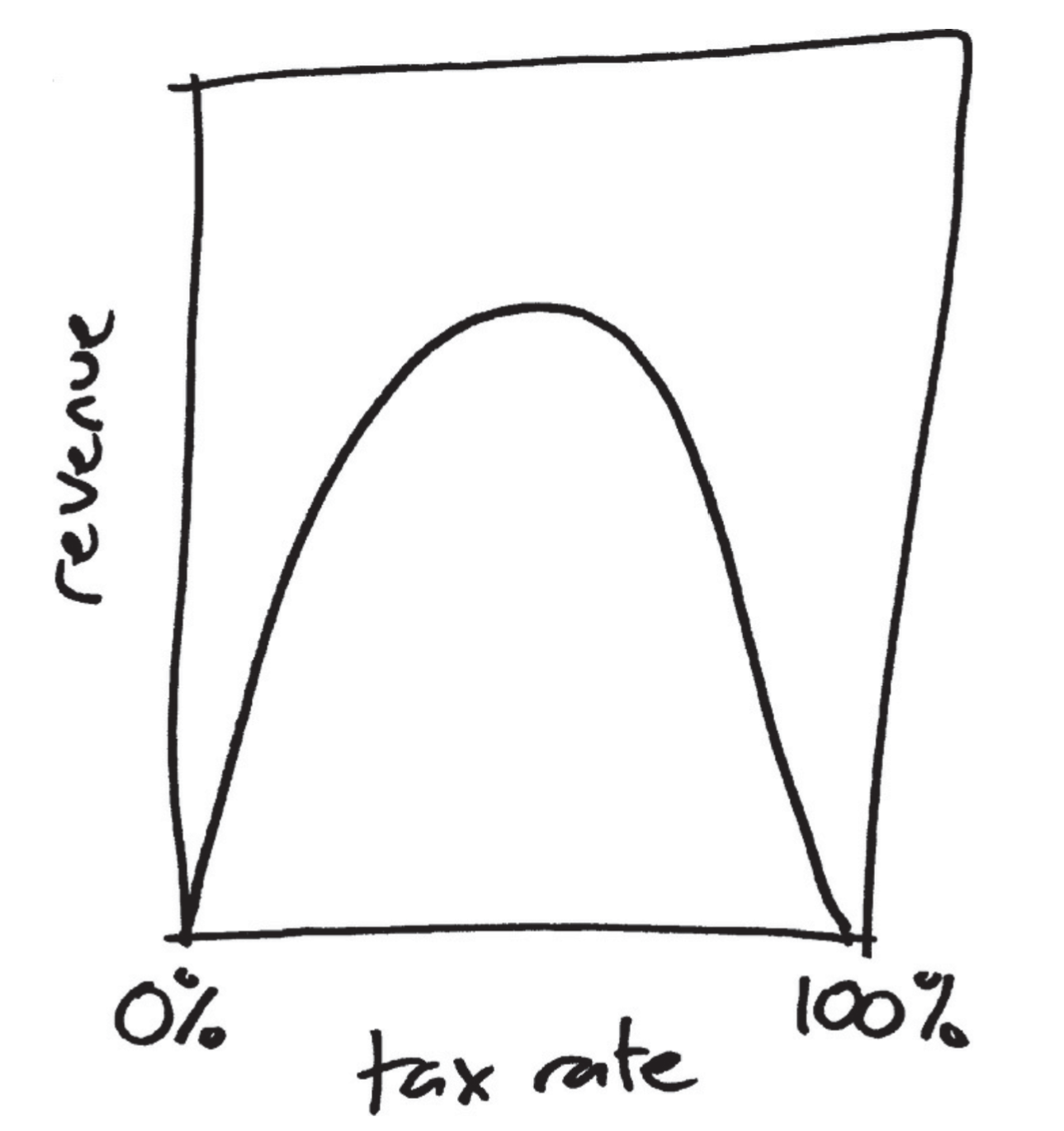
The legend of the Laffer curve goes like this: Arthur Laffer, then an economics professor at the University of Chicago, had dinner one night in 1974 with Dick Cheney, Donald Rumsfeld, and Wall Street Journal editor Jude Wanniski at an upscale hotel restaurant in Washington DC. They were tussling over President Ford’s tax plan, and eventually, as intellectuals do when the tussling gets heavy, Laffer commandeered a napkin and drew a picture. The picture looked like this:
\texttt{sales} \approx \beta_0 + \beta_1 \times \texttt{TV}
\texttt{sales} \approx \beta_0 + \beta_1 \times \texttt{TV}+ \beta_2 \times \texttt{radio}
We’ll focus on one predictor and one response variable for most of this lecture.
Using a term like nonlinear science is like referring to the bulk of zoology as the study of non-elephant animals. (Stanisław Ulam)
Download a file called Mx_1x1.txt from the Human Mortality Database.
No-one is allowed to distribute the data, but you can download it for free. Here are the first few rows to get a sense of what it looks like.
with open('Mx_1x1.txt', 'r') as file:
for i in range(10):
print(file.readline(), end='')Luxembourg, Death rates (period 1x1), Last modified: 09 Aug 2023; Methods Protocol: v6 (2017)
Year Age Female Male Total
1960 0 0.023863 0.039607 0.031891
1960 1 0.001690 0.003528 0.002644
1960 2 0.001706 0.002354 0.002044
1960 3 0.001257 0.002029 0.001649
1960 4 0.000844 0.001255 0.001051
1960 5 0.000873 0.001701 0.001293
1960 6 0.000443 0.000430 0.000437lux <- read_table("Mx_1x1.txt", skip = 2, show_col_types = FALSE) %>%
rename(age=Age, year=Year, mx=Female) %>%
select(age, year, mx) %>%
filter(age != '110+') %>%
mutate(year = as.integer(year), age = as.integer(age), mx = as.numeric(mx))lux# A tibble: 6,930 × 3
age year mx
<int> <int> <dbl>
1 0 1960 0.0239
2 1 1960 0.00169
3 2 1960 0.00171
4 3 1960 0.00126
5 4 1960 0.000844
6 5 1960 0.000873
7 6 1960 0.000443
8 7 1960 0
9 8 1960 0.000951
10 9 1960 0
# ℹ 6,920 more rowssummary(lux) age year mx
Min. : 0.0 Min. :1960 Min. :0.00000
1st Qu.: 27.0 1st Qu.:1975 1st Qu.:0.00045
Median : 54.5 Median :1991 Median :0.00337
Mean : 54.5 Mean :1991 Mean :0.09200
3rd Qu.: 82.0 3rd Qu.:2007 3rd Qu.:0.04178
Max. :109.0 Max. :2022 Max. :6.00000
NA's :358 lux = pd.read_csv('Mx_1x1.txt', sep='\s+', skiprows=2)
lux = lux.rename(columns={'Age': 'age', 'Year': 'year', 'Female': 'mx'})
lux = lux[['age', 'year', 'mx']]
lux = lux[lux['age'] != '110+'].copy()
lux.loc[lux['mx'] == '.', 'mx'] = np.nan
lux['year'] = lux['year'].astype(int)
lux['age'] = lux['age'].astype(int)
lux['mx'] = lux['mx'].astype(float)
lux age year mx
0 0 1960 0.023863
1 1 1960 0.001690
2 2 1960 0.001706
3 3 1960 0.001257
4 4 1960 0.000844
... ... ... ...
6987 105 2022 0.661481
6988 106 2022 5.419704
6989 107 2022 NaN
6990 108 2022 NaN
6991 109 2022 NaN
[6930 rows x 3 columns]average_df <- lux %>% group_by(age) %>% summarise(mx = mean(mx, na.rm = TRUE))
ggplot(lux, aes(x = age, y = mx)) +
geom_line(aes(group = year, color = year), alpha = 0.3) +
geom_line(data = average_df, color = "black", linewidth = 1) +
labs(x = "Age", y = "Mortality Rate", color = "Year") +
theme_minimal()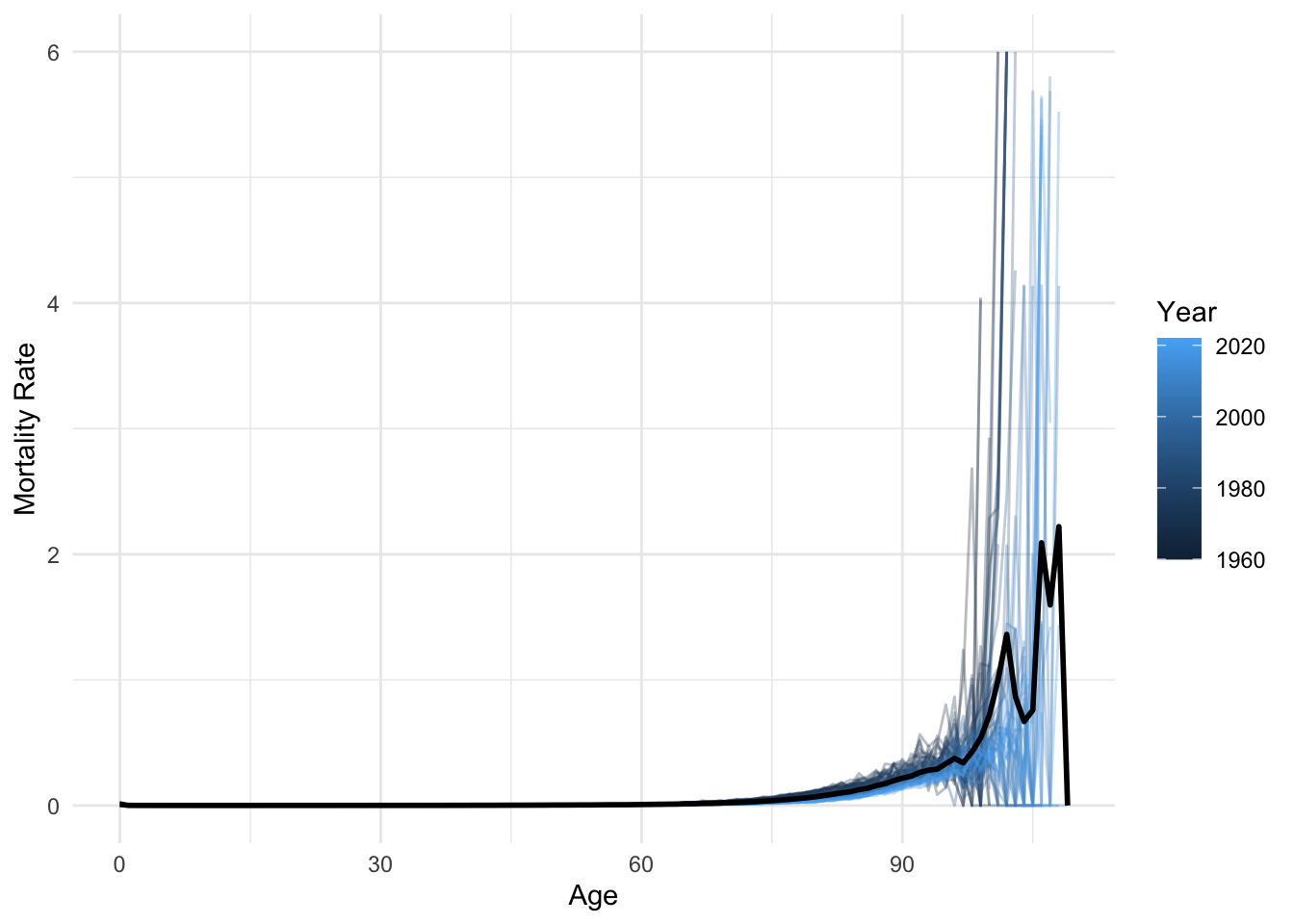
average_df = lux.groupby('age')[['mx']].mean().reset_index()
sns.lineplot(data=lux, x='age', y='mx', hue='year', palette='viridis', alpha=0.3)
plt.plot(average_df['age'], average_df['mx'], color='black', linewidth=2)
plt.xlabel('Age'); plt.ylabel('Mortality Rate');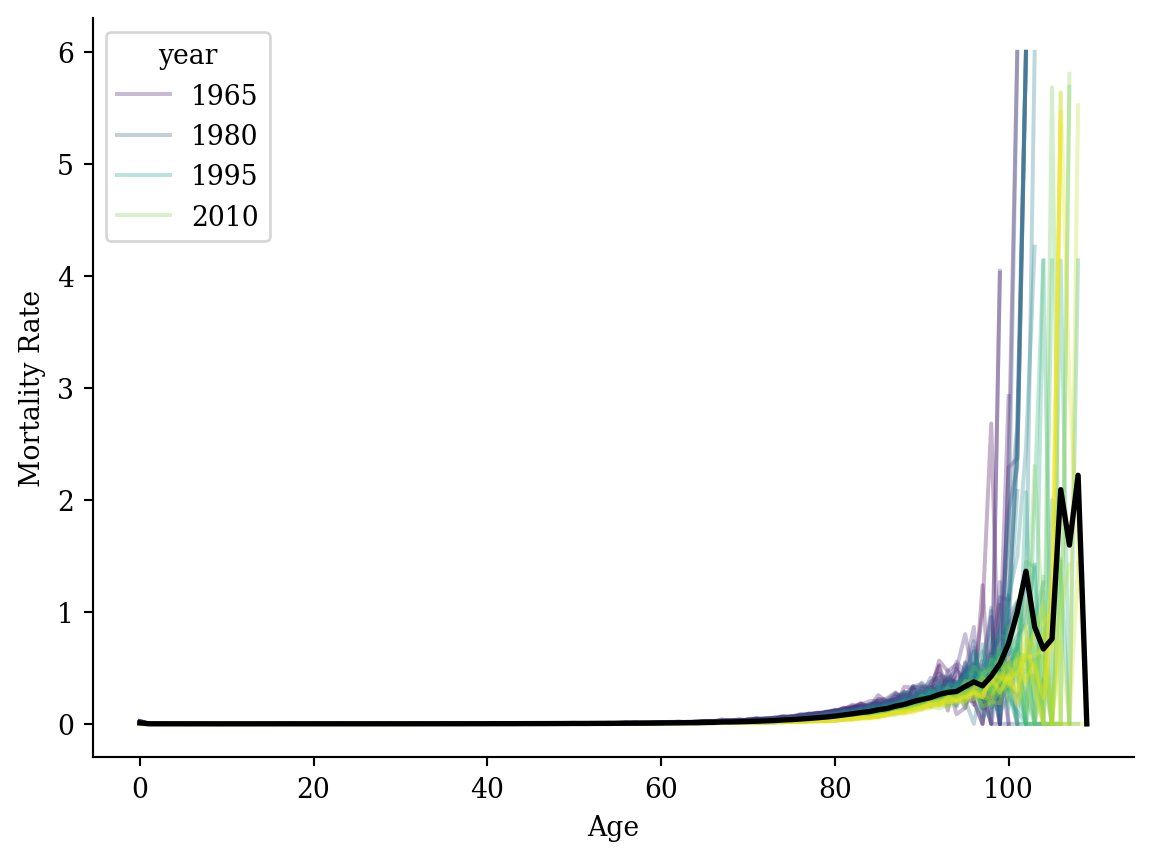
lux <- lux %>% filter(age <= 90)average_df <- average_df %>% filter(age <= 90)
ggplot(lux, aes(x = age, y = mx)) +
geom_line(aes(group = year, color = year), alpha = 0.3) +
geom_line(data = average_df, color = "black", linewidth = 1) +
labs(x = "Age", y = "Mortality Rate", color = "Year") +
theme_minimal()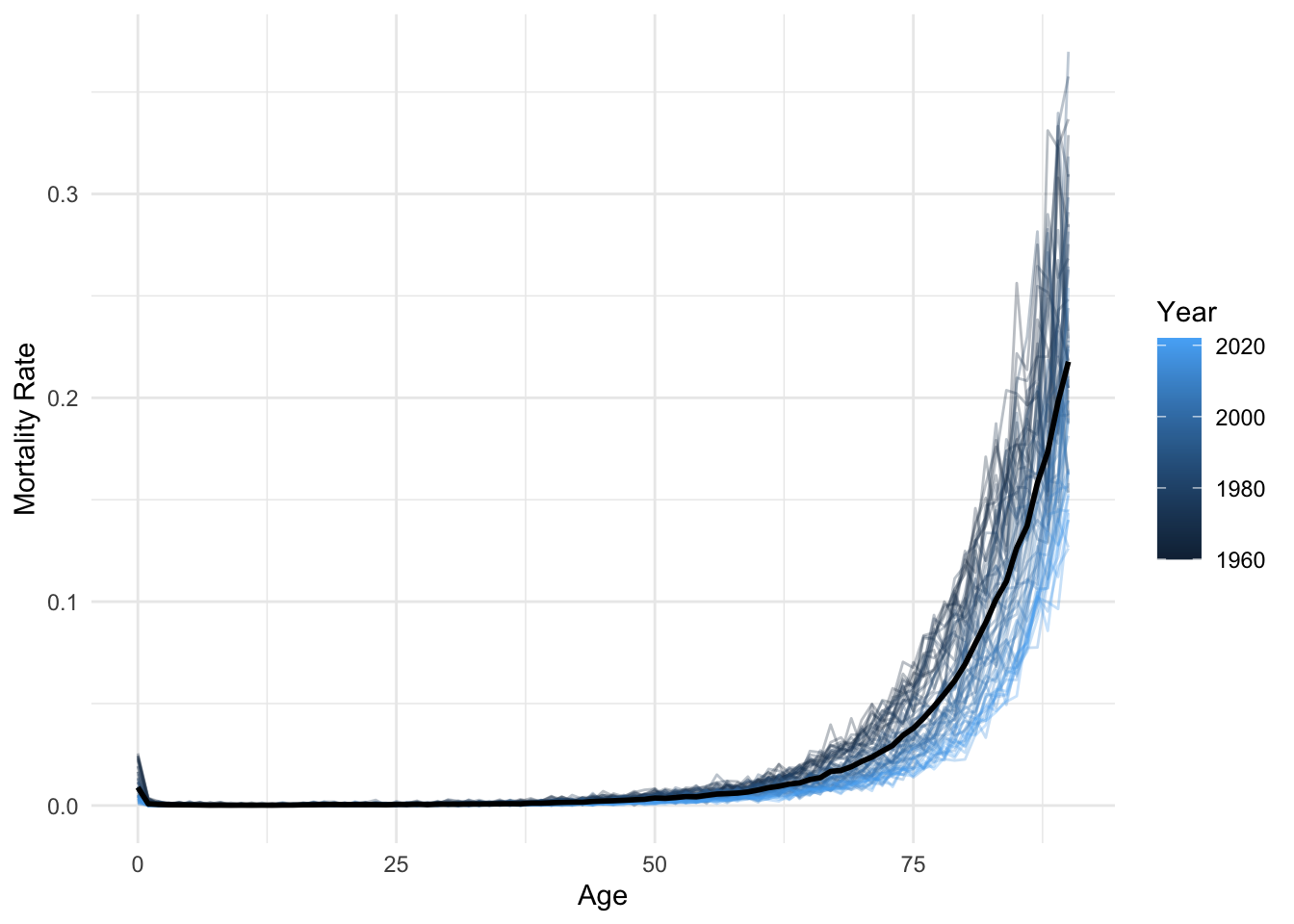
lux = lux[lux['age'] <= 90].copy()average_df = average_df[average_df['age'] <= 90]
sns.lineplot(data=lux, x='age', y='mx', hue='year', palette='viridis', alpha=0.3)
plt.plot(average_df['age'], average_df['mx'], color='black', linewidth=2)
plt.xlabel('Age'); plt.ylabel('Mortality Rate');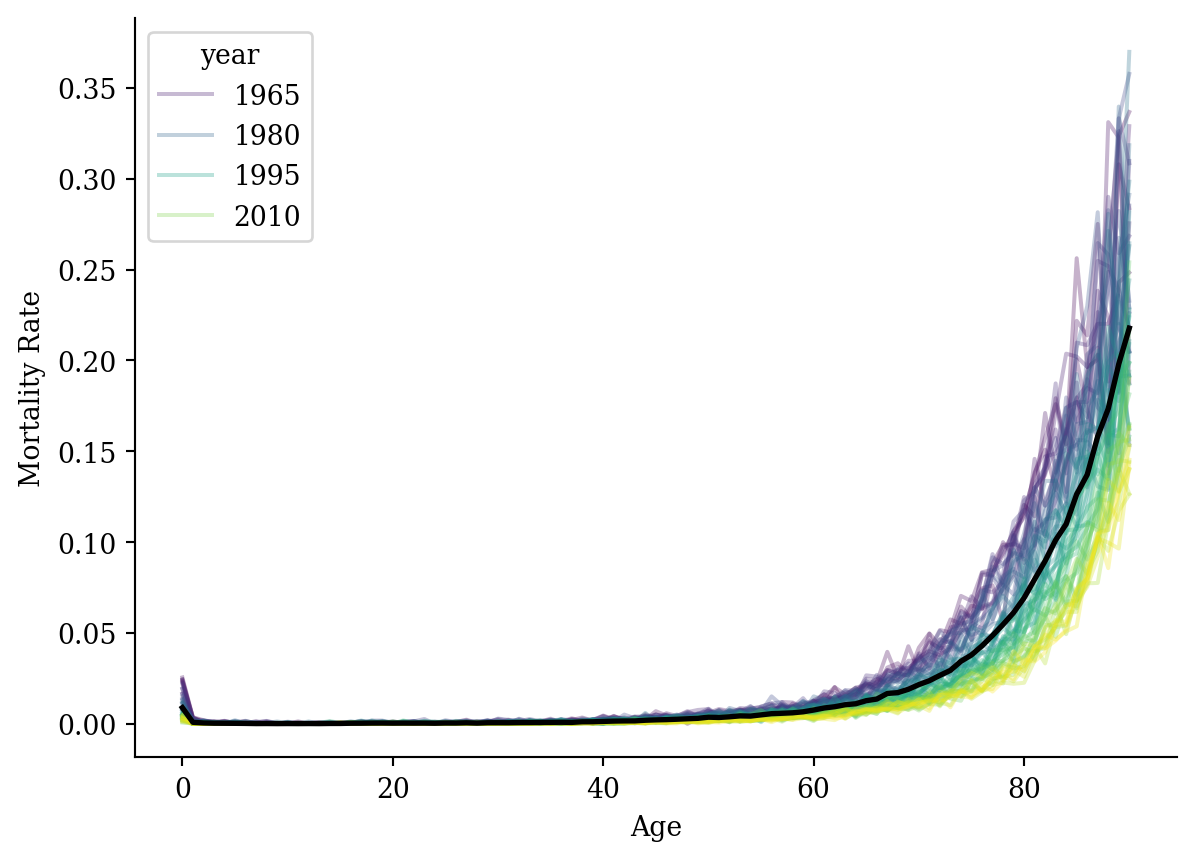
lux$log_mx <- log(lux$mx)
lux <- lux[lux$log_mx != -Inf, ]average_df <- average_df %>%
left_join(lux %>%
group_by(age) %>%
summarise(log_mx = mean(log_mx, na.rm = TRUE)),
by = "age")
ggplot(lux, aes(x = age, y = log_mx)) +
geom_line(aes(group = year, color = year), alpha = 0.3) +
geom_line(data = average_df, color = "black", linewidth = 1) +
labs(x = "Age", y = "Log-Mortality Rate", color = "Year") +
theme_minimal()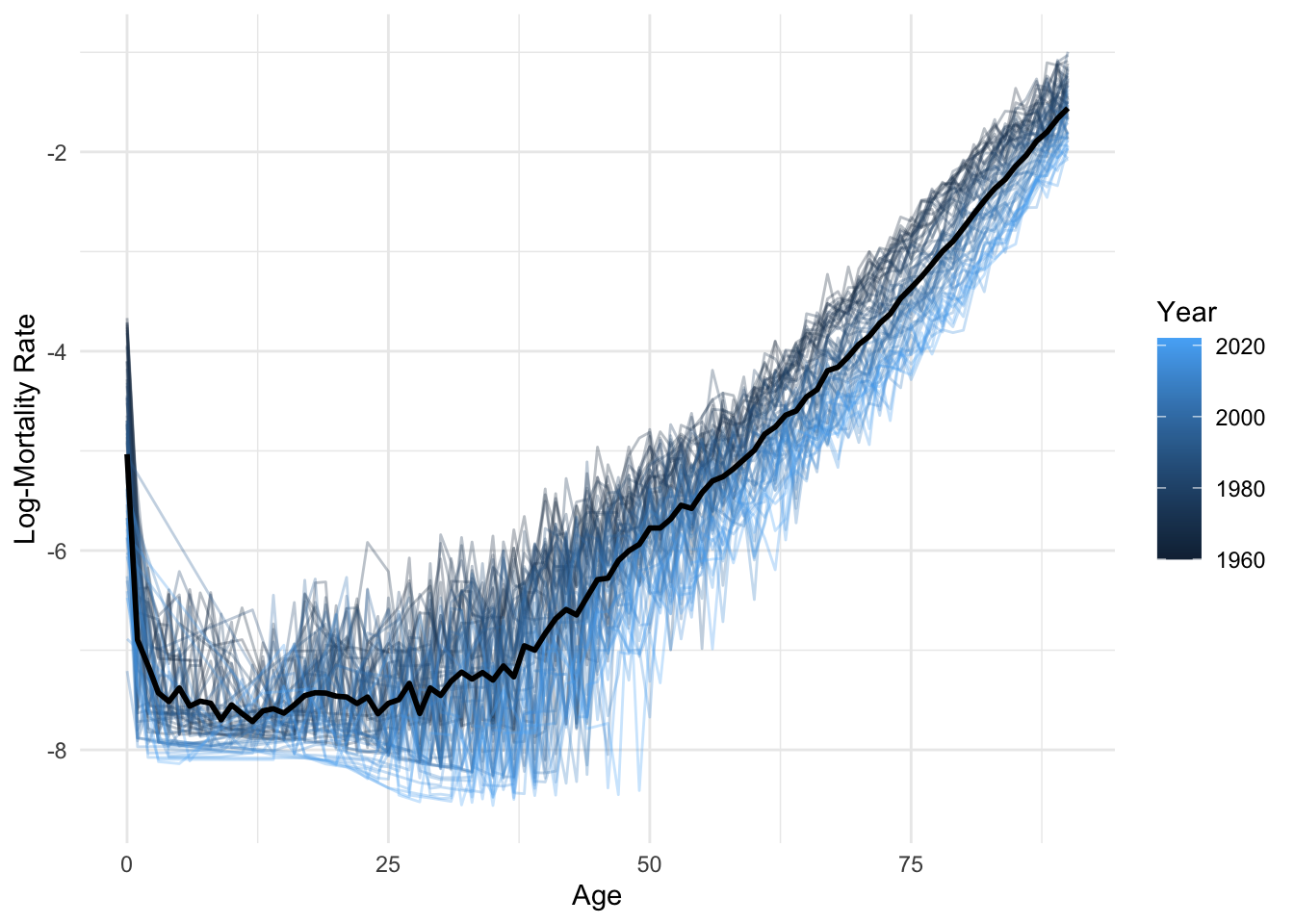
lux['log_mx'] = np.log(lux['mx'])C:\Users\Patrick\MAMBAF~1\envs\ml2025\Lib\site-packages\pandas\core\arraylike.py:399: RuntimeWarning: divide by zero encountered in log
result = getattr(ufunc, method)(*inputs, **kwargs)lux = lux[lux['log_mx'] != -np.inf]average_df['log_mx'] = lux.groupby('age')[['log_mx']].mean()
sns.lineplot(data=lux, x='age', y='log_mx', hue='year', palette='viridis', alpha=0.3)
plt.plot(average_df['age'], average_df['log_mx'], color='black', linewidth=2)
plt.xlabel('Age'); plt.ylabel('Log-Mortality Rate');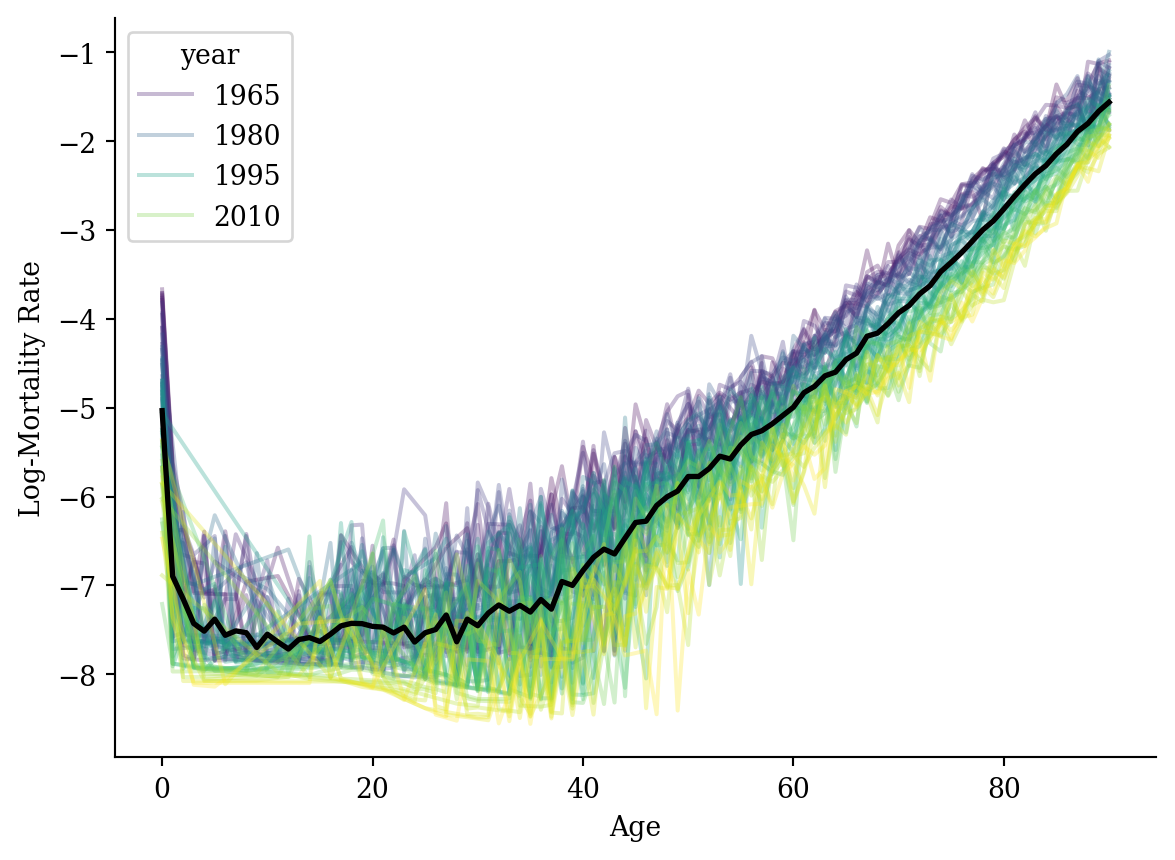
lux_2020 <- lux %>% filter(year == 2020)
model_lr <- lm(log_mx ~ age, data = lux_2020)p <- ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
geom_point(alpha = 0.75) + labs(x = "Age", y = "Log-Mortality")
age_grid <- data.frame(age = seq(min(lux$age), max(lux$age), by=0.1))
predictions <- predict(model_lr, newdata = age_grid)
p + geom_line(data = age_grid, aes(x = age, y = predictions),
color = "red", linewidth=2)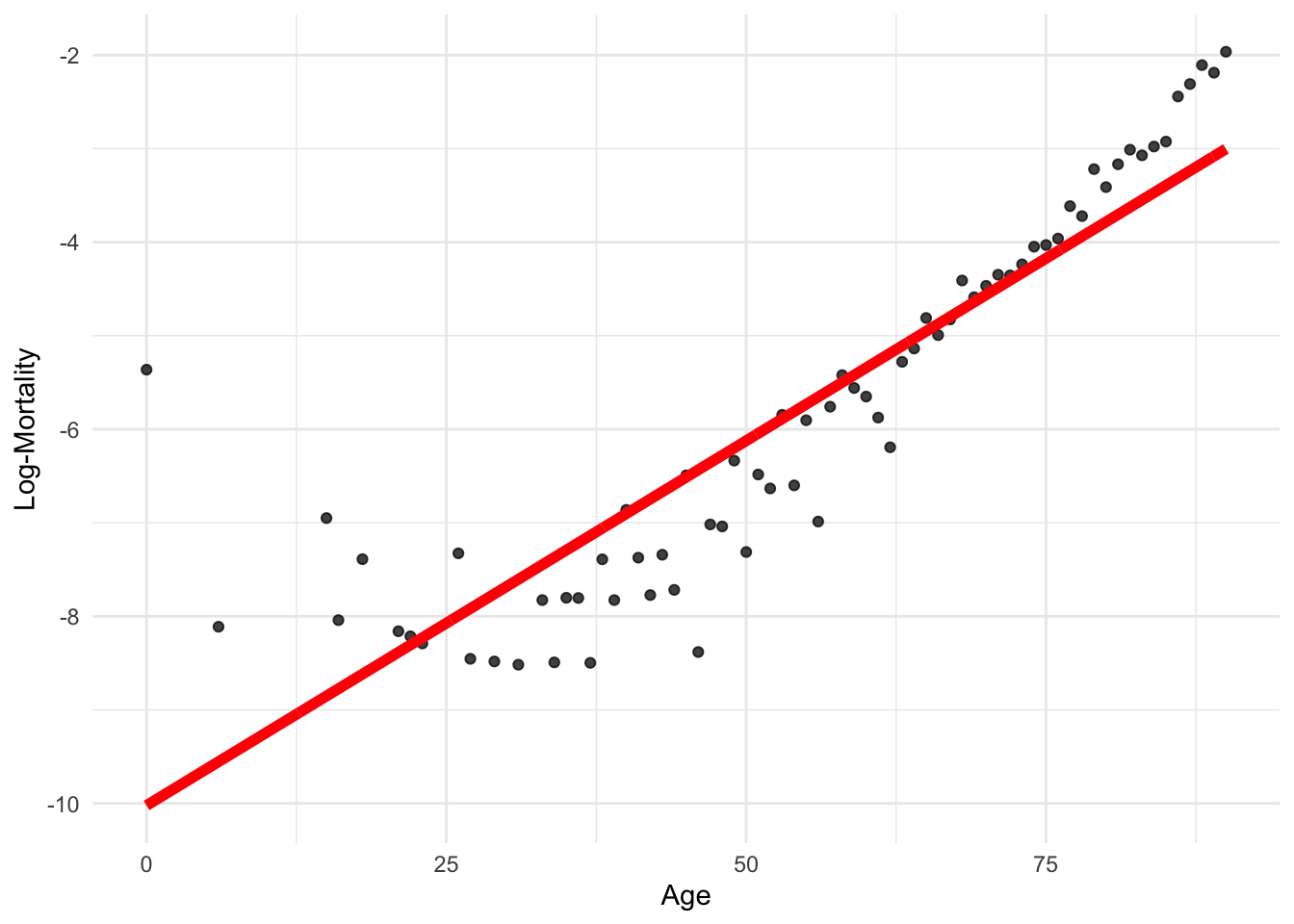
lux_2020 = lux[lux['year'] == 2020].copy()
model_lr = smf.ols('log_mx ~ age', data=lux_2020).fit()plt.scatter(lux_2020['age'], lux_2020['log_mx'], alpha=0.75)
plt.plot(lux_2020['age'], model_lr.predict(lux_2020[['age']]), color='red')
plt.xlabel('Age'); plt.ylabel('Log-Mortality');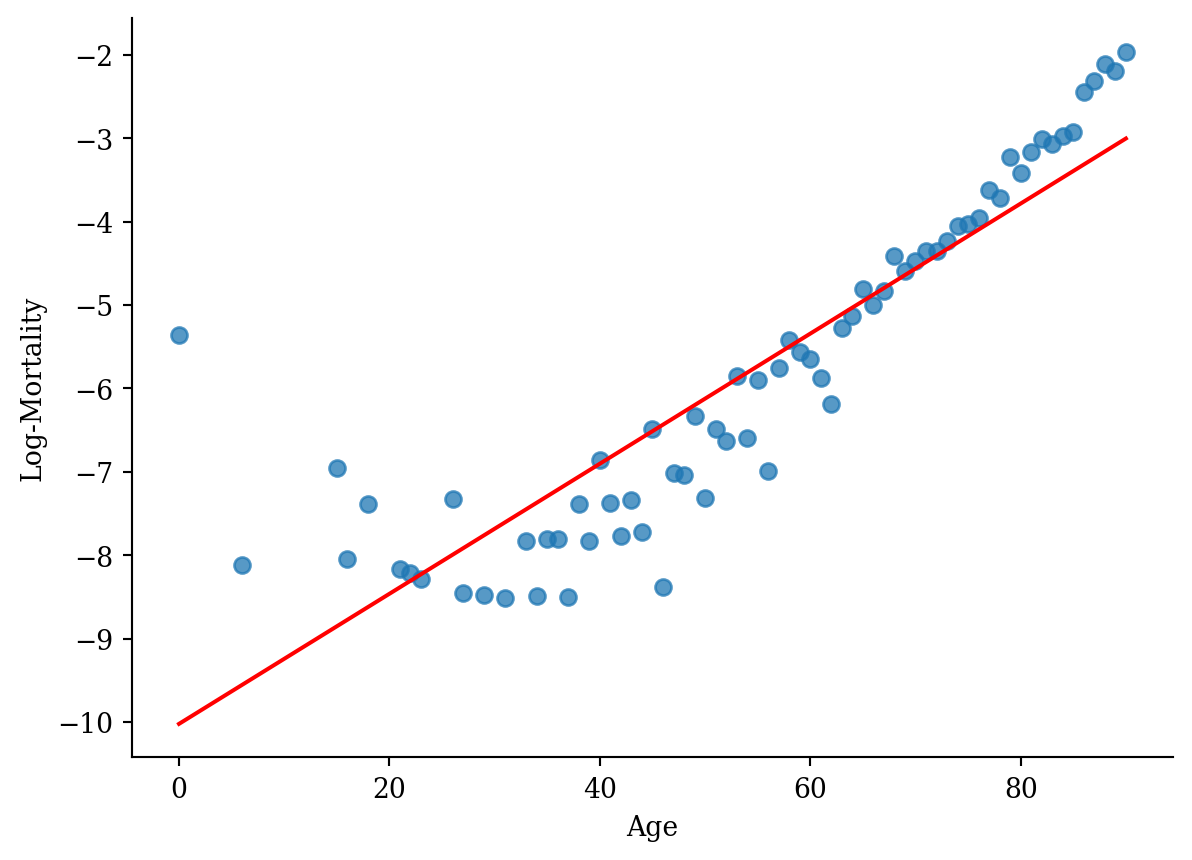
model_quad <- lm(log_mx ~ poly(age, 2), data = lux_2020)predictions <- predict(model_quad, newdata = age_grid)
p + geom_line(data = age_grid, aes(x = age, y = predictions),
color = "red", linewidth=2)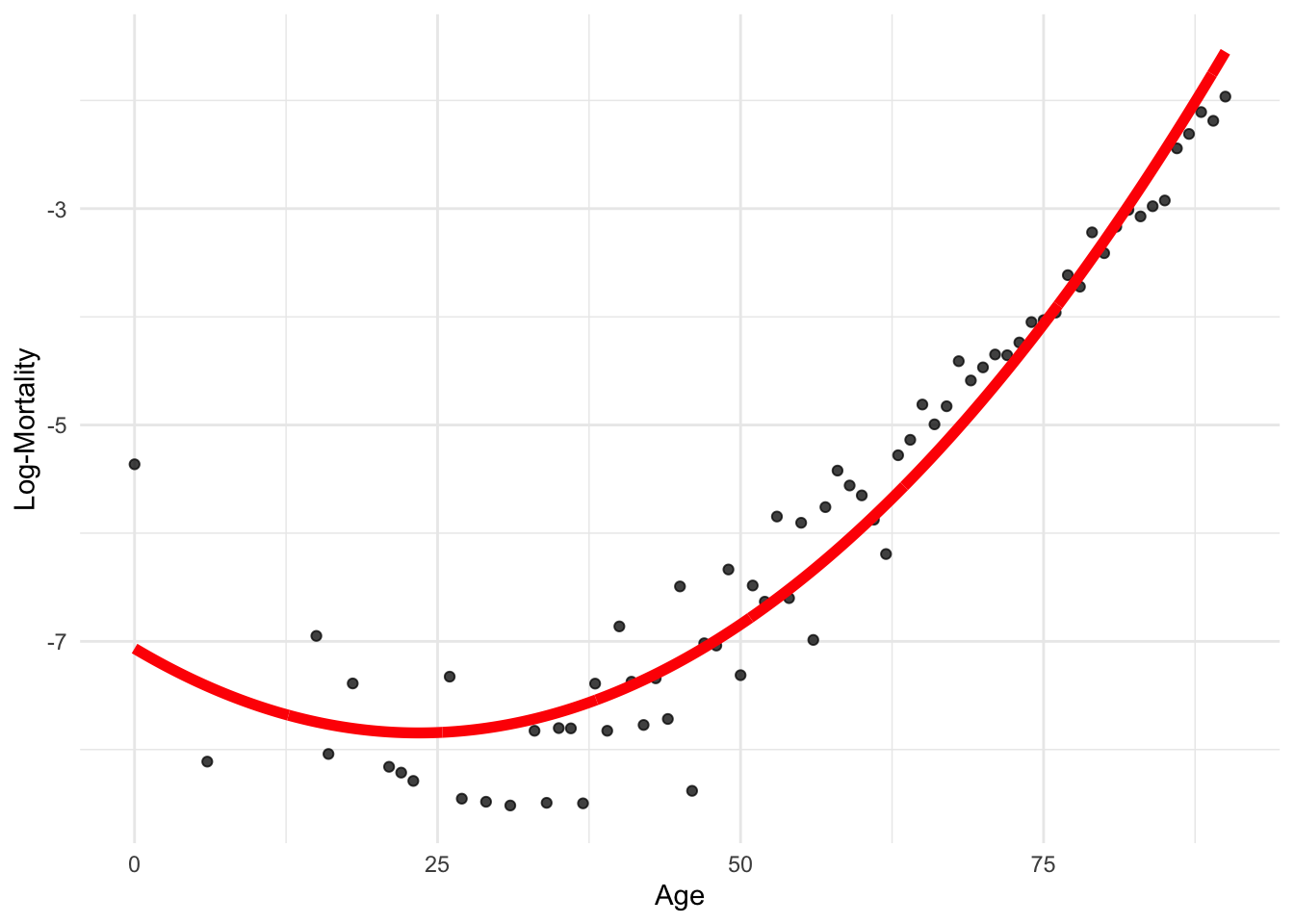
model_quad = smf.ols('log_mx ~ np.power(age, 2) + age', data=lux_2020).fit()plt.scatter(lux_2020['age'], lux_2020['log_mx'], alpha=0.75)
plt.plot(lux_2020['age'], model_quad.predict(lux_2020), color='red', linewidth=2)
plt.xlabel('Age'); plt.ylabel('Log Mortality');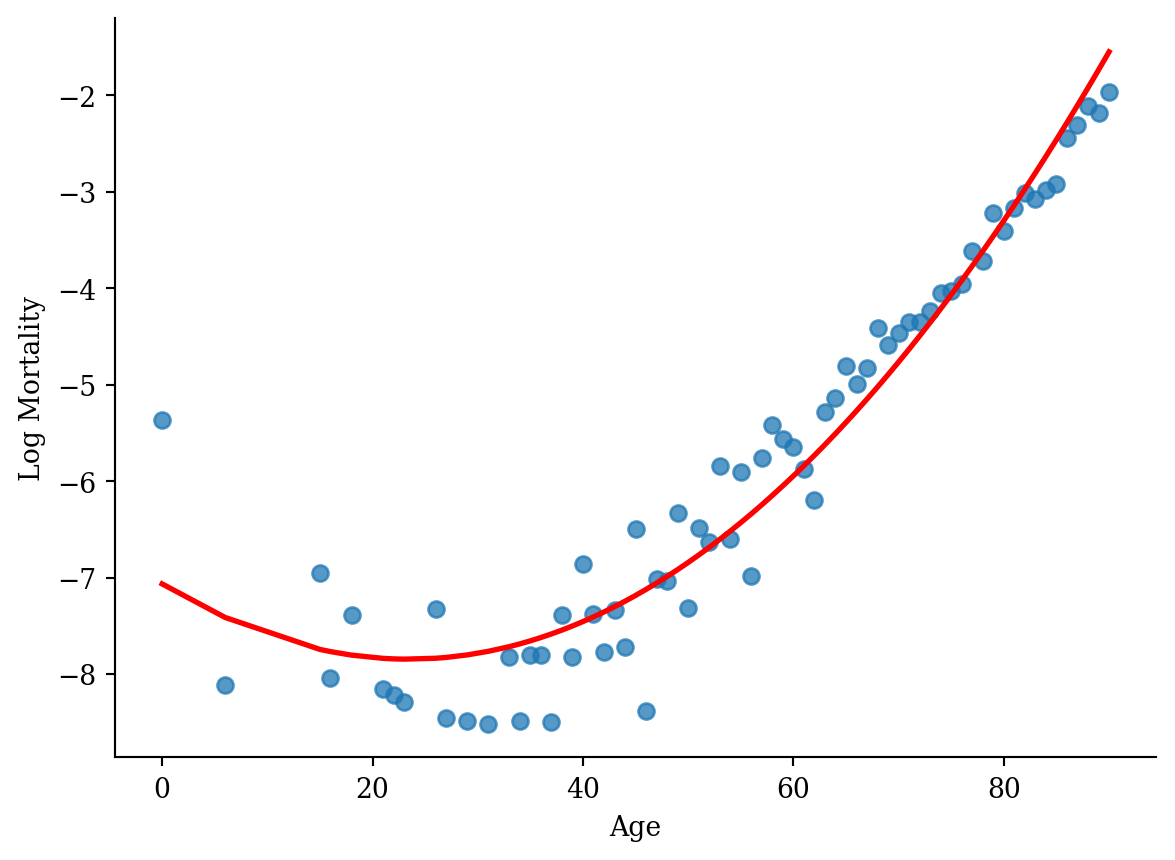
model_step <- lm(log_mx ~ cut(age, seq(0, 90, 10), right=F), data = lux_2020)p <- ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
geom_point(alpha = 0.75) + labs(x = "Age", y = "Log-Mortality")
age_grid <- data.frame(age = seq(min(lux$age), max(lux$age), by=0.1))
predictions <- predict(model_step, newdata = age_grid)
p + geom_point(data = age_grid, aes(x = age, y = predictions),
color = "red", size=2)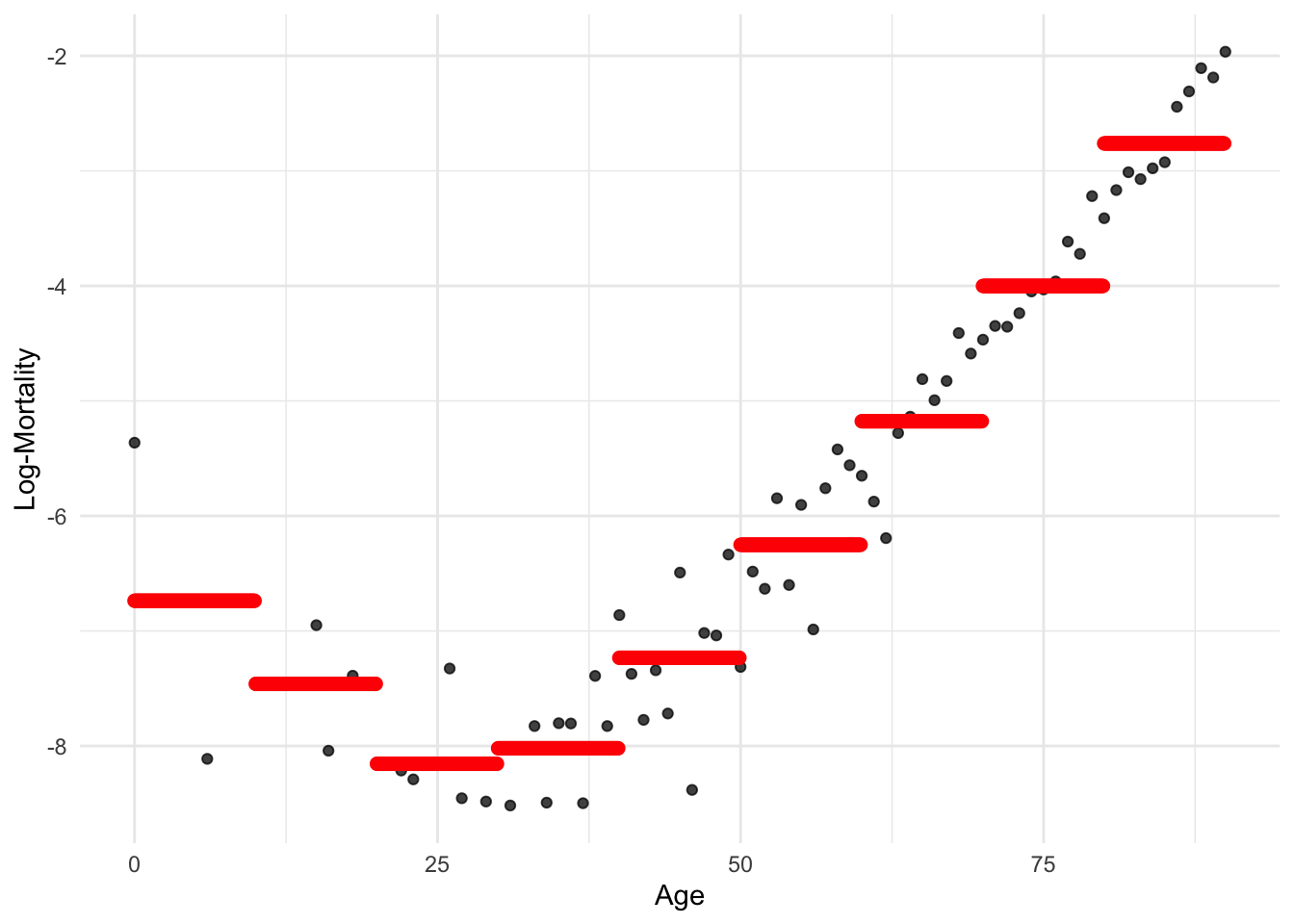
lux_2020['age_bin'] = pd.cut(lux_2020['age'], bins=np.arange(0, 101, 10), right=False)
lux_2020['age_bin_str'] = lux_2020['age_bin'].astype(str)
model_step = smf.ols('log_mx ~ age_bin_str', data=lux_2020).fit()# Generate a DataFrame for predictions
# This involves creating a DataFrame where 'age' spans the unique bins used in the model
age_grid = np.linspace(0, 101, 1000)
df_plot = pd.DataFrame({
'age': age_grid,
'age_bin_str': pd.cut(age_grid, bins=np.arange(0, 101, 10), right=False).astype(str)}
)
# Predict using the model
df_plot['predictions'] = model_step.predict(df_plot)
# Plot
plt.scatter(lux_2020['age'], lux_2020['log_mx'], alpha=0.75)
plt.scatter(df_plot['age'], df_plot['predictions'], color='red', linewidth=2)
plt.xlabel('Age')
plt.ylabel('Log-Mortality')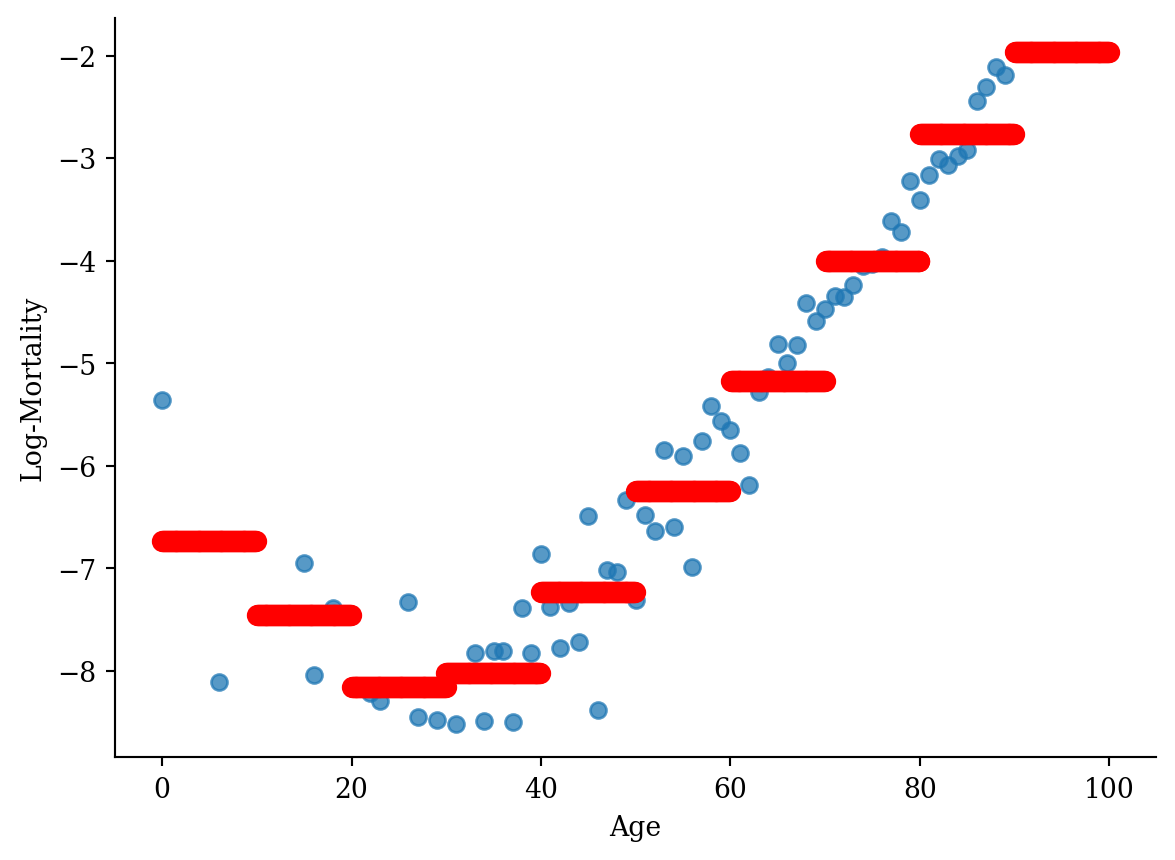
model_spline <- lm(log_mx ~ bs(age, degree=10), data=lux_2020) # Requires splines packagepredictions <- predict(model_spline, newdata = age_grid)
p + geom_line(data = age_grid, aes(x = age, y = predictions),
color = "red", linewidth=2, alpha=0.75)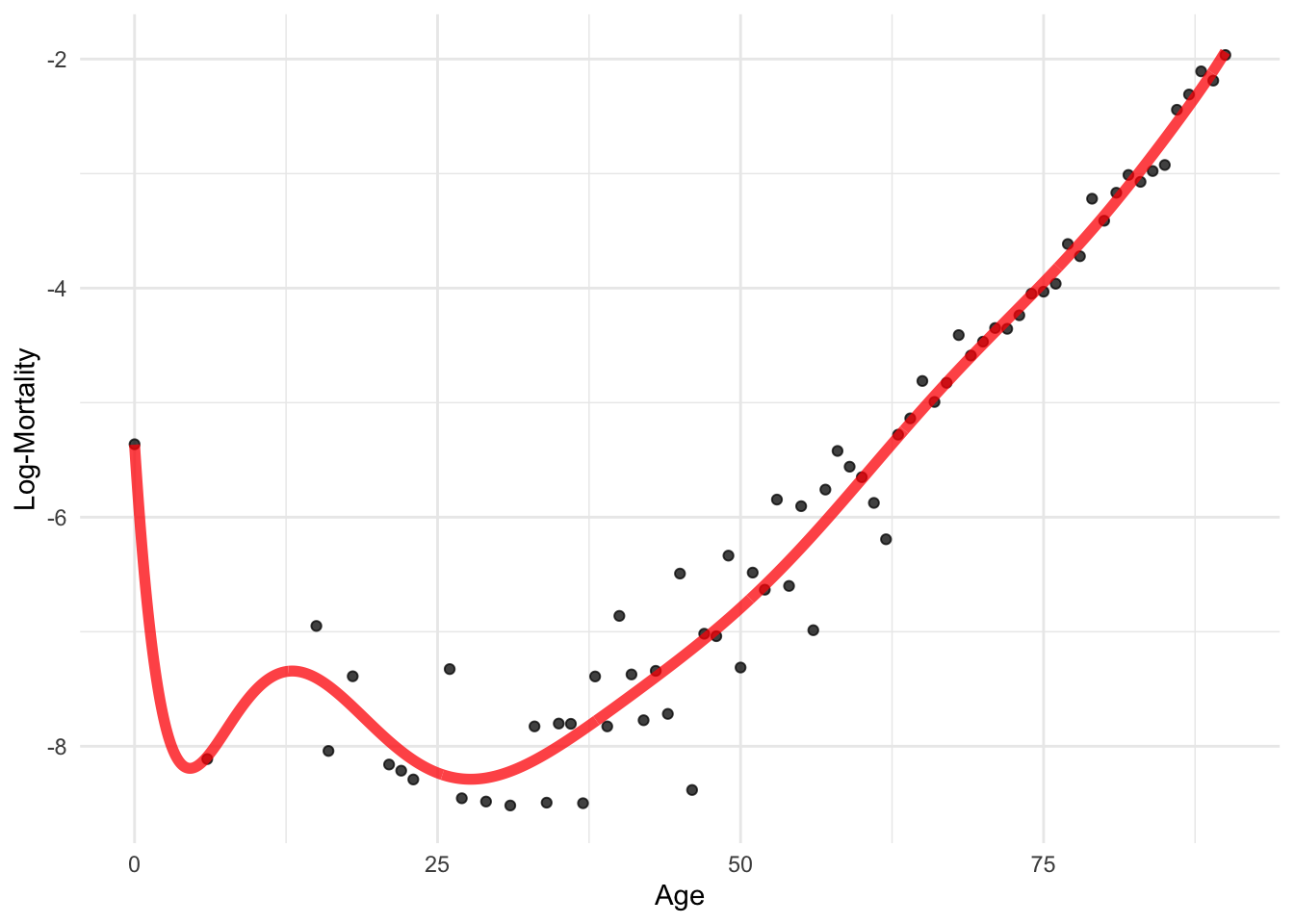
spline_x = dmatrix("bs(lux_2020['age'], degree=3, knots=[15,30,45])",
{"lux_2020['age']": lux_2020['age']}, return_type='dataframe')
model_spline = sm.GLM(lux_2020['log_mx'], spline_x).fit()# Prepare data for plotting
x_range = pd.DataFrame({'age': np.linspace(lux_2020['age'].min(), lux_2020['age'].max(), 100)})
x_range_transformed = dmatrix("bs(x_range, degree=3, knots=[15,30,45])", {"x_range": x_range['age']}, return_type='dataframe')
predictions_spline = model_spline.predict(x_range_transformed)
plt.scatter(lux_2020['age'], lux_2020['log_mx'], alpha=0.75)
plt.plot(x_range['age'], predictions_spline, color='red')
plt.xlabel('Age'); plt.ylabel('Log Mortality');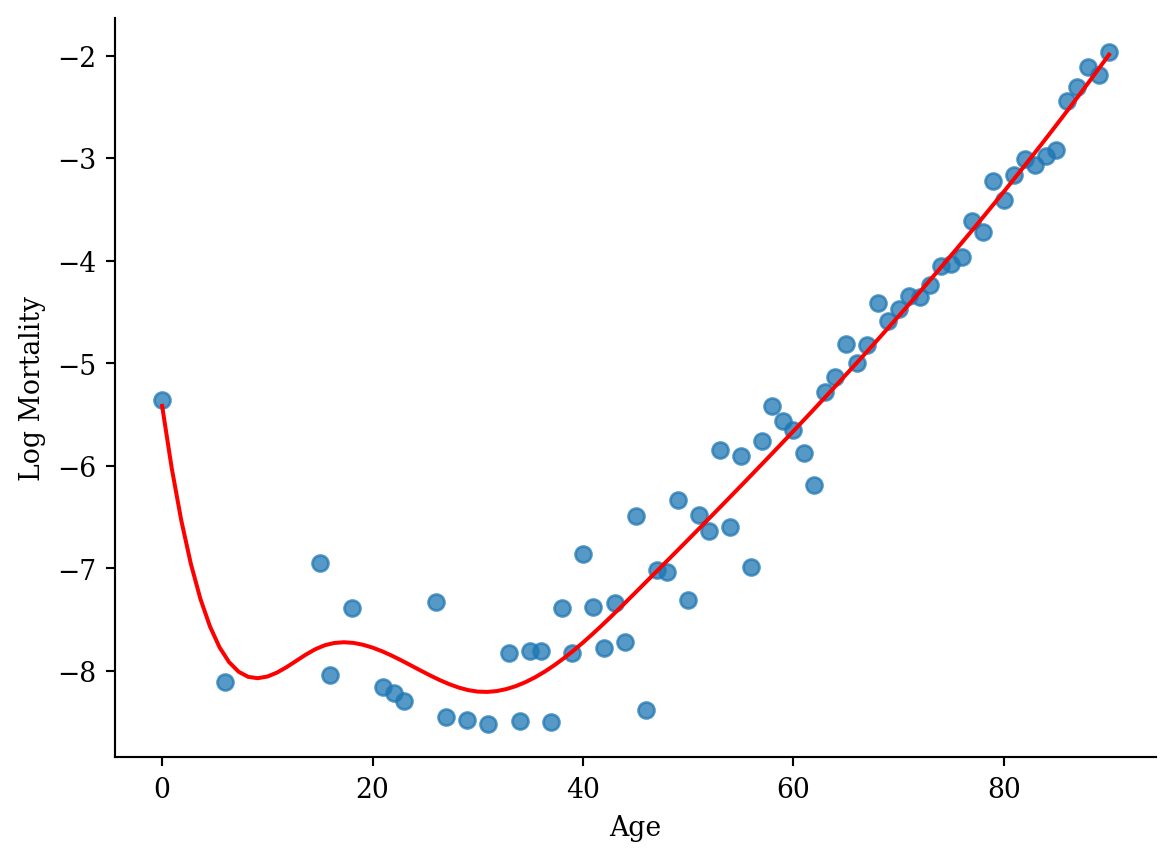
Methods from this class (p. 8–9):
Take ACTL3141/ACTL5104 for mortality modelling, ACTL3143/ACTL5111 for actuarial AI
ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
geom_point(aes(y = predict(model_lr)), color = "red", size = 2) +
geom_point(alpha = 0.75, size = 2) + labs(x = "Age", y = "Log-Mortality")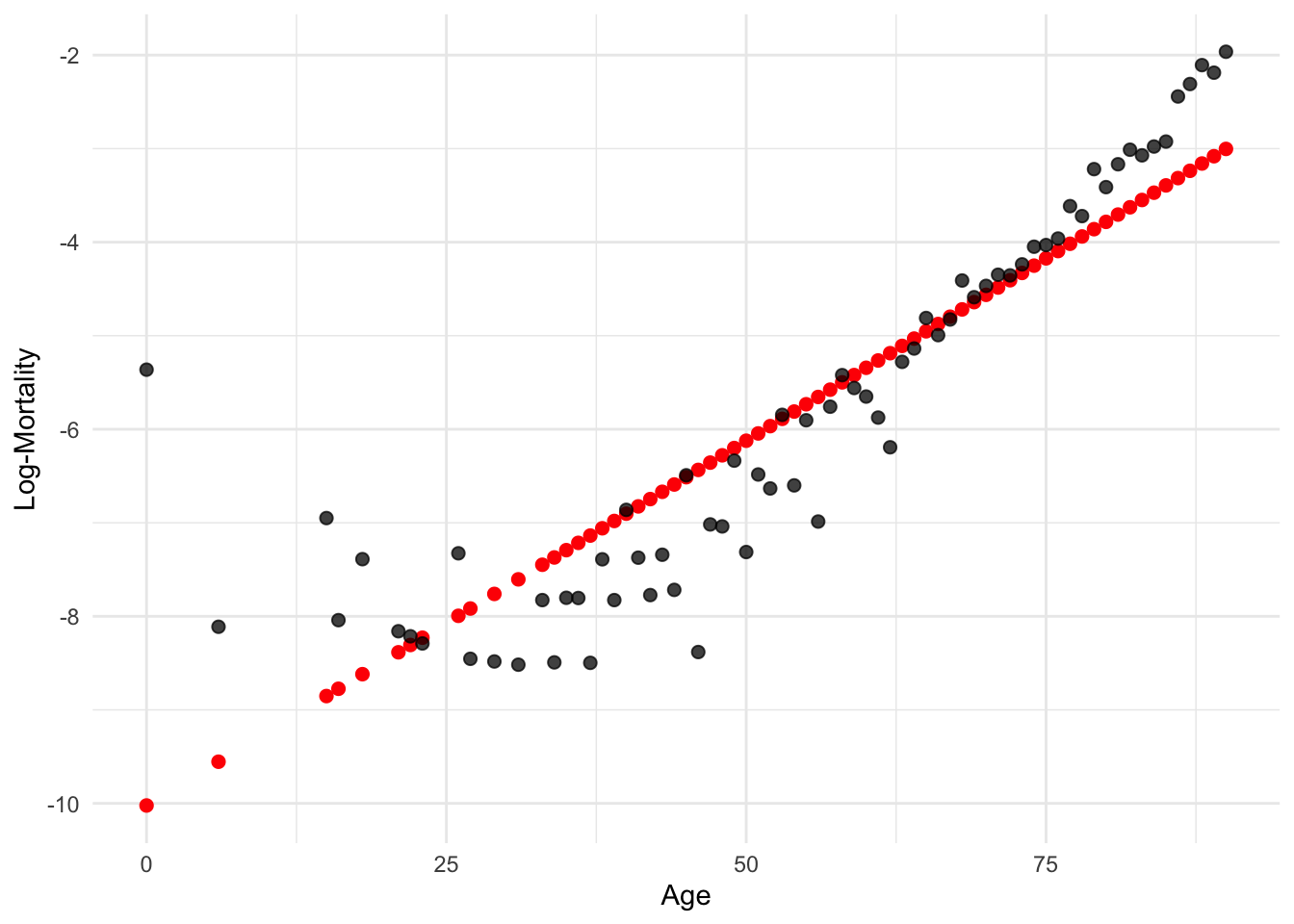
model = smf.ols('log_mx ~ age', data=lux_2020).fit()
plt.scatter(lux_2020['age'], model.predict(lux_2020[['age']]), color='red')
plt.scatter(lux_2020['age'], lux_2020['log_mx'], alpha=0.75)
plt.xlabel('Age'); plt.ylabel('Log-Mortality');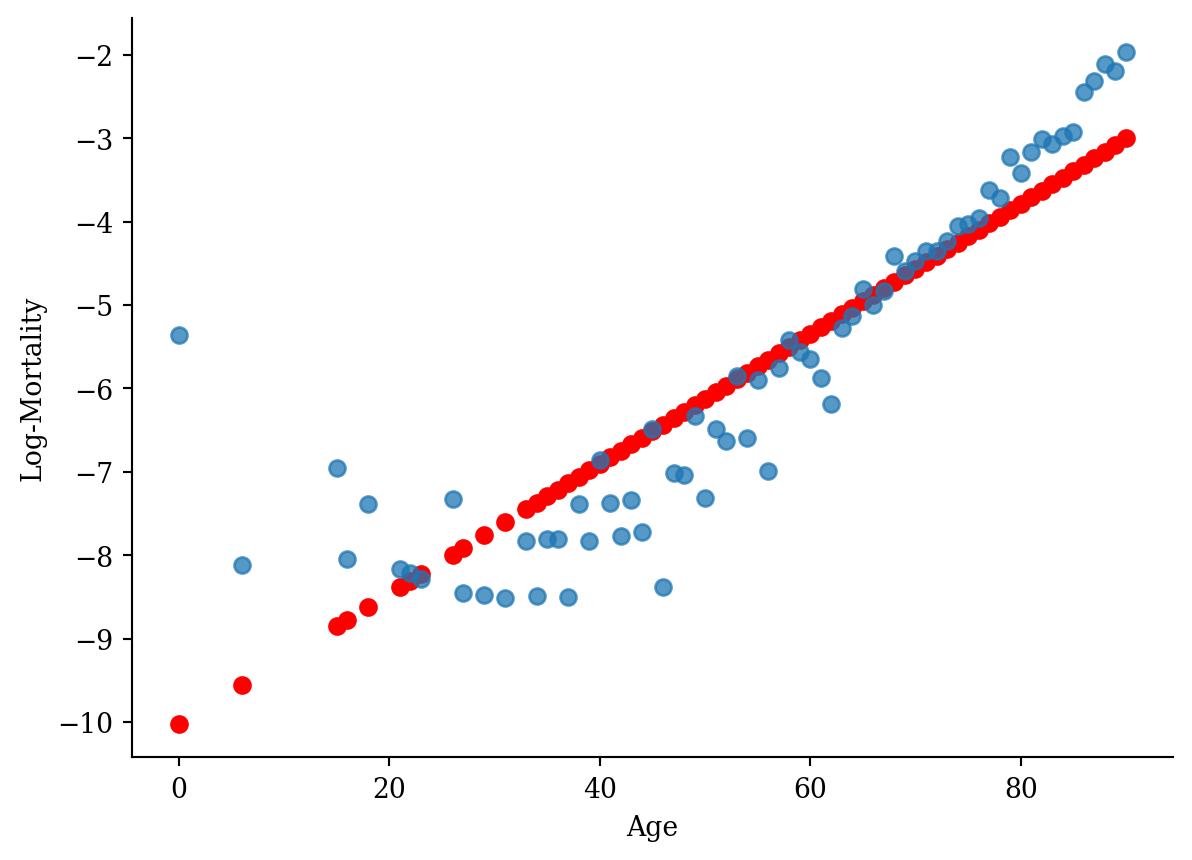
ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
geom_smooth(method = "lm", formula = y ~ x, color = "red", linewidth=2) +
geom_point(alpha = 0.75) + labs(x = "Age", y = "Log Mortality")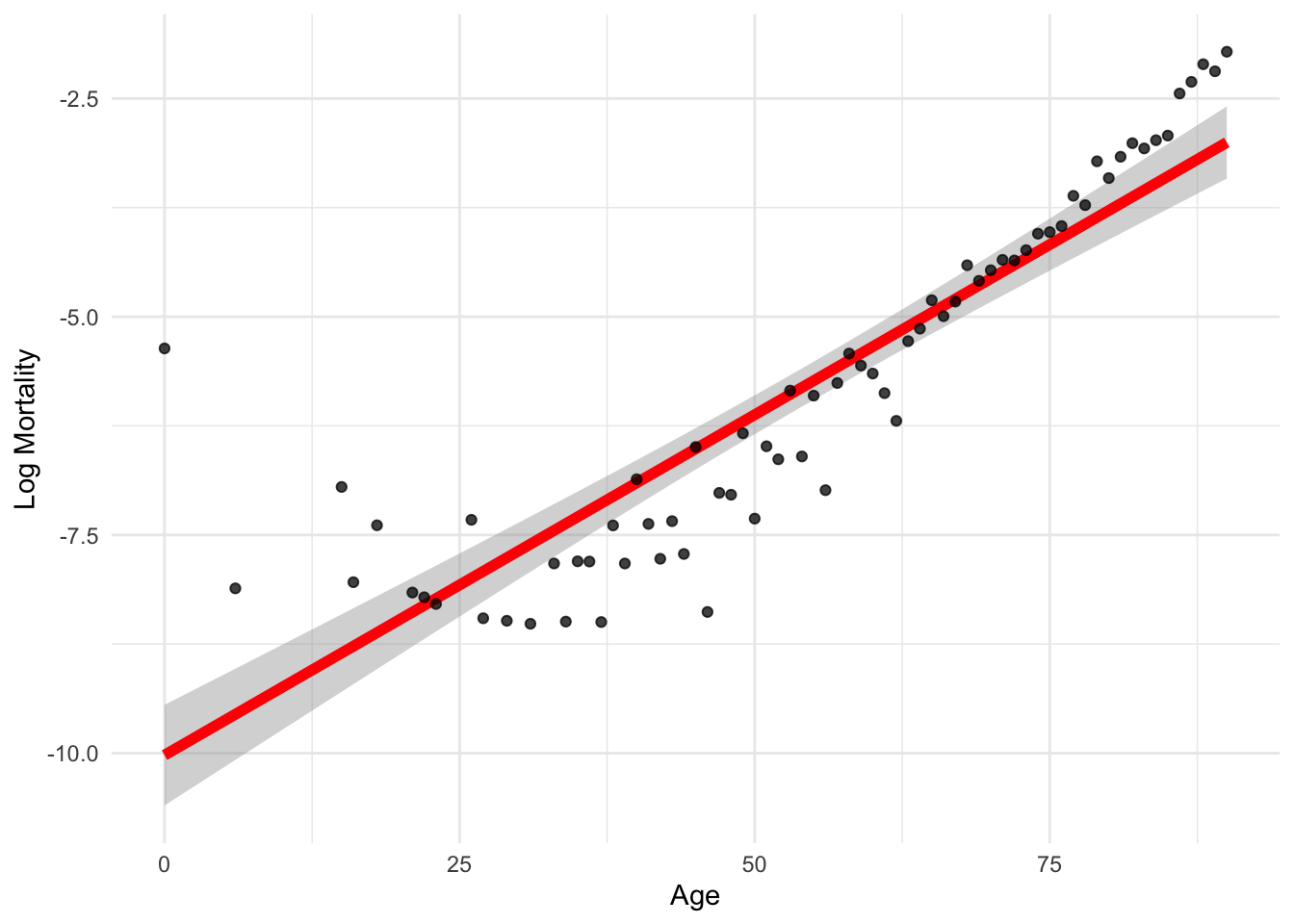
sns.regplot(x='age', y='log_mx', data=lux_2020, ci=95)
plt.xlabel('Age'); plt.ylabel('Log-Mortality');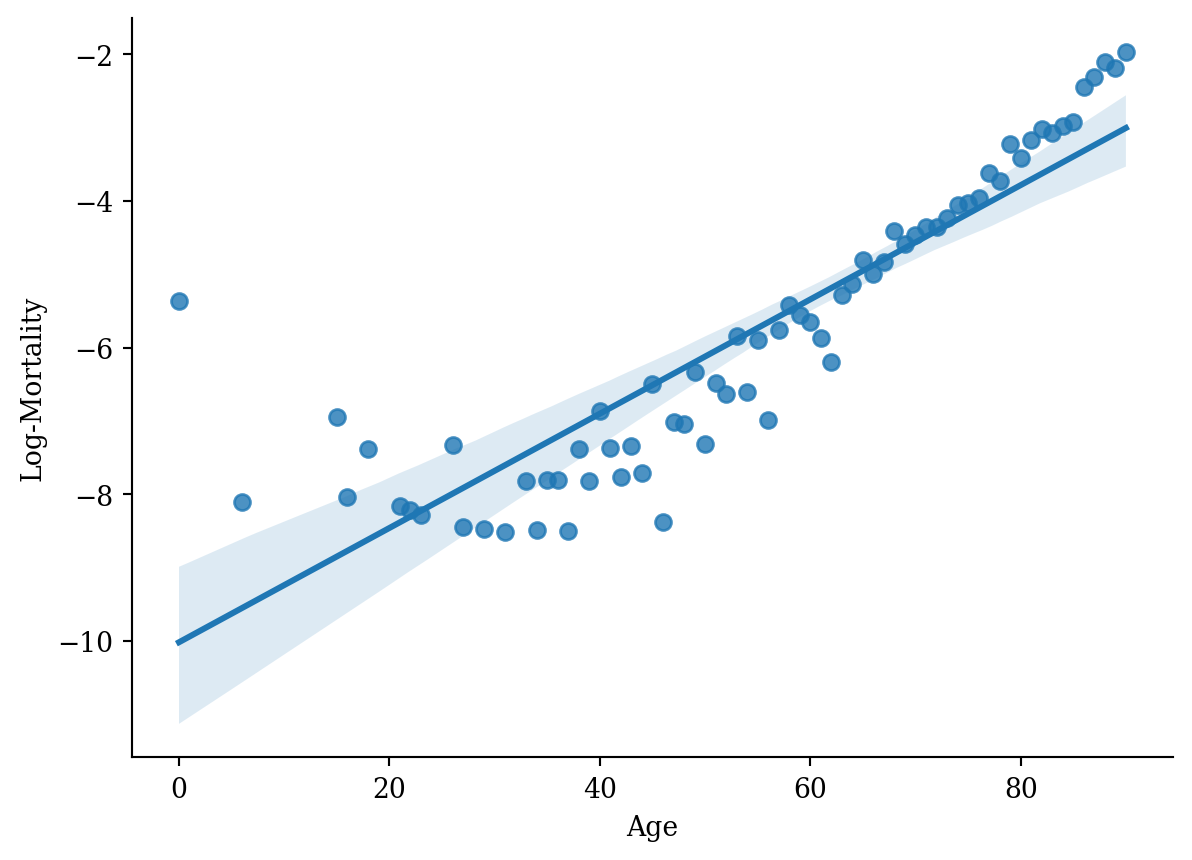
df_grid <- data.frame(age = seq(25, 35, by = 0.5))
df_grid$log_mx <- predict(model_lr, newdata = df_grid)lux_young <- lux_2020 %>% filter(age >= 25, age <= 35)
ggplot(lux_young, aes(x = age, y = log_mx)) +
theme_minimal() +
geom_point(data = df_grid, aes(y = log_mx), color = "red", size = 2) +
geom_point(alpha = 0.75, size = 2) +
labs(x = "Age", y = "Log-Mortality")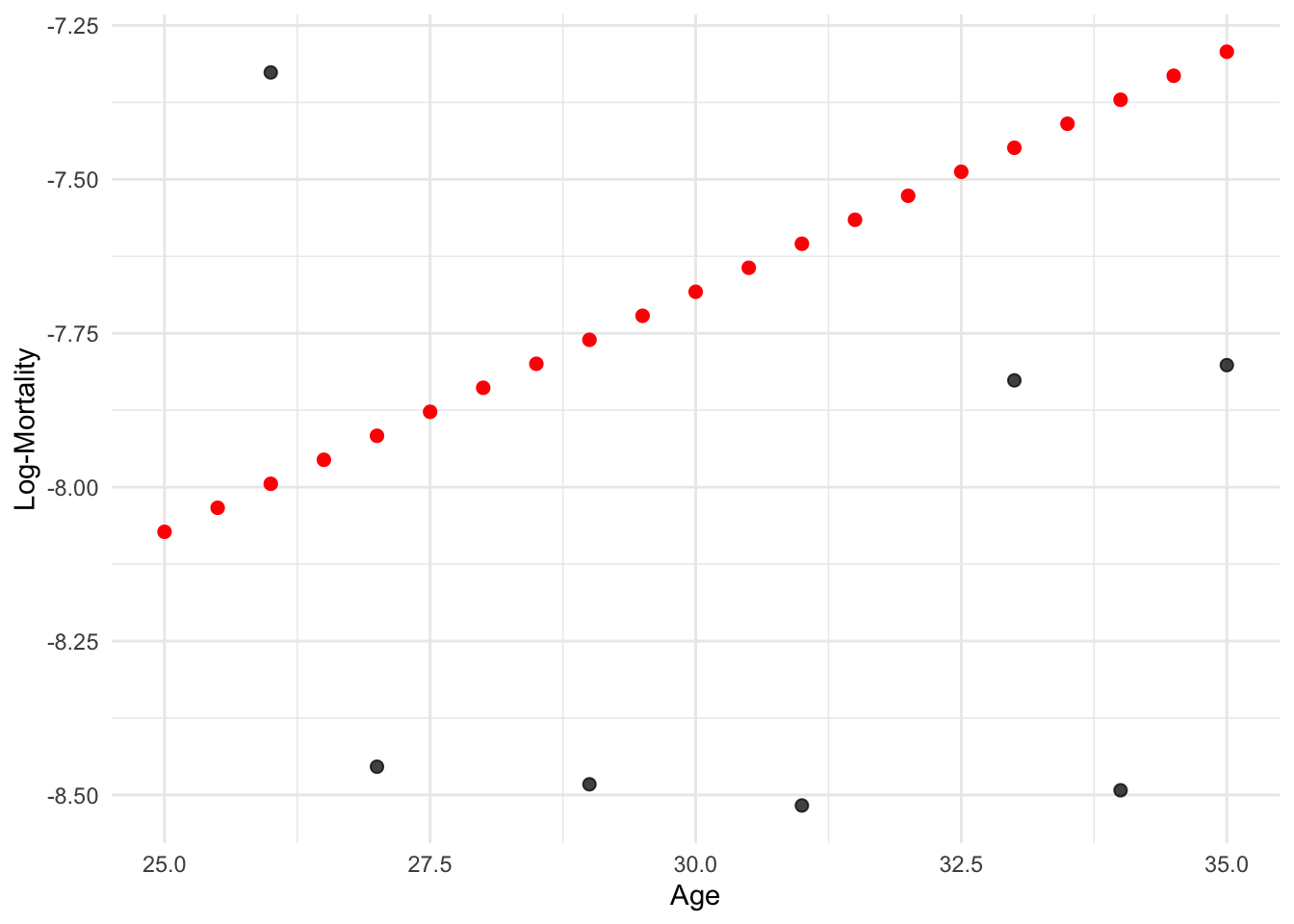
age_grid = pd.DataFrame({"age": np.arange(25, 35, 0.5)})
predictions = model.predict(age_grid)lux_young = lux_2020[(lux_2020['age'] >= 25) & (lux_2020['age'] <= 35)]
plt.scatter(lux_young['age'], lux_young['log_mx'], alpha=0.75)
plt.scatter(age_grid, model.predict(age_grid), color='red', marker='o')
plt.xlabel('Age'); plt.ylabel('Log-Mortality');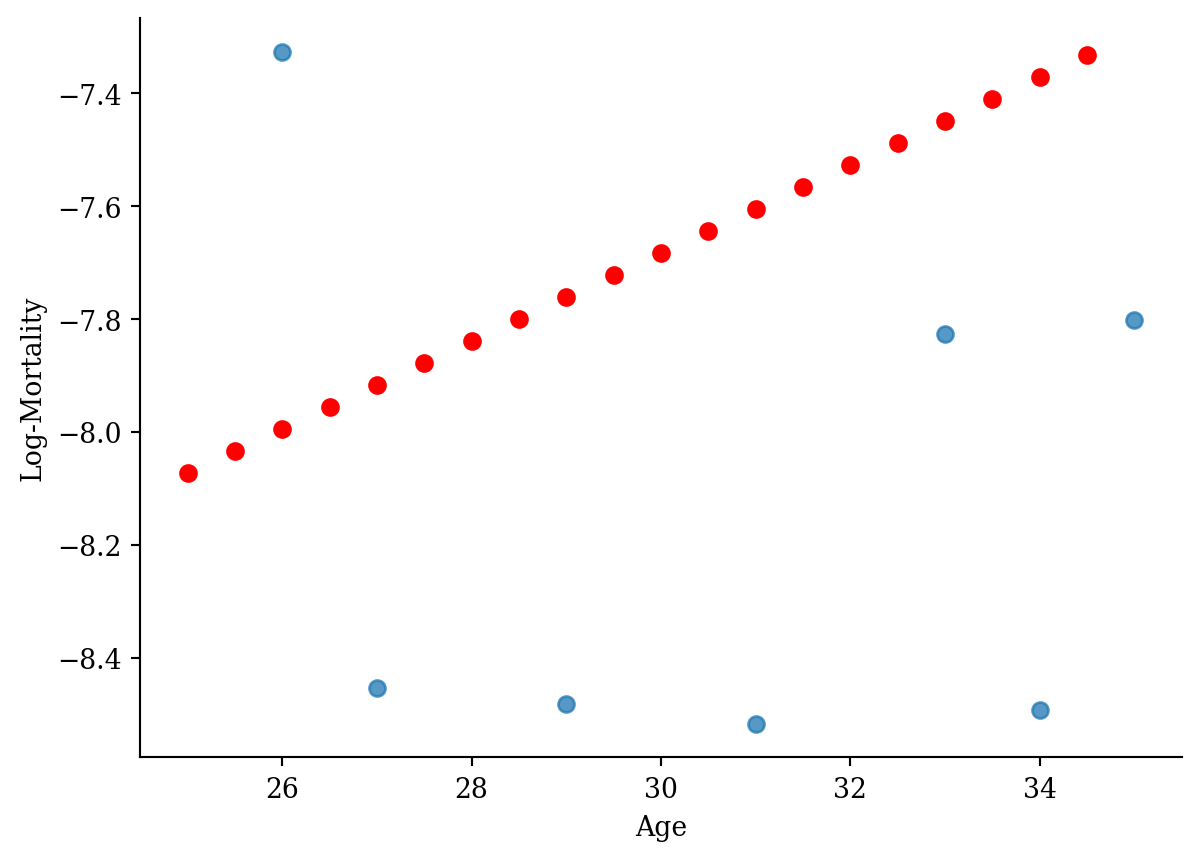
df_grid <- data.frame(age = seq(40, 130))
df_grid$log_mx <- predict(model_lr, newdata = df_grid)lux_old <- lux_2020 %>% filter(age >= 40, age <= 130)
ggplot(lux_old, aes(x = age, y = log_mx)) +
theme_minimal() +
geom_line(data = df_grid, aes(y = log_mx), color = "red", linewidth = 2) +
geom_point(alpha = 0.75) +
labs(x = "Age", y = "Log-Mortality")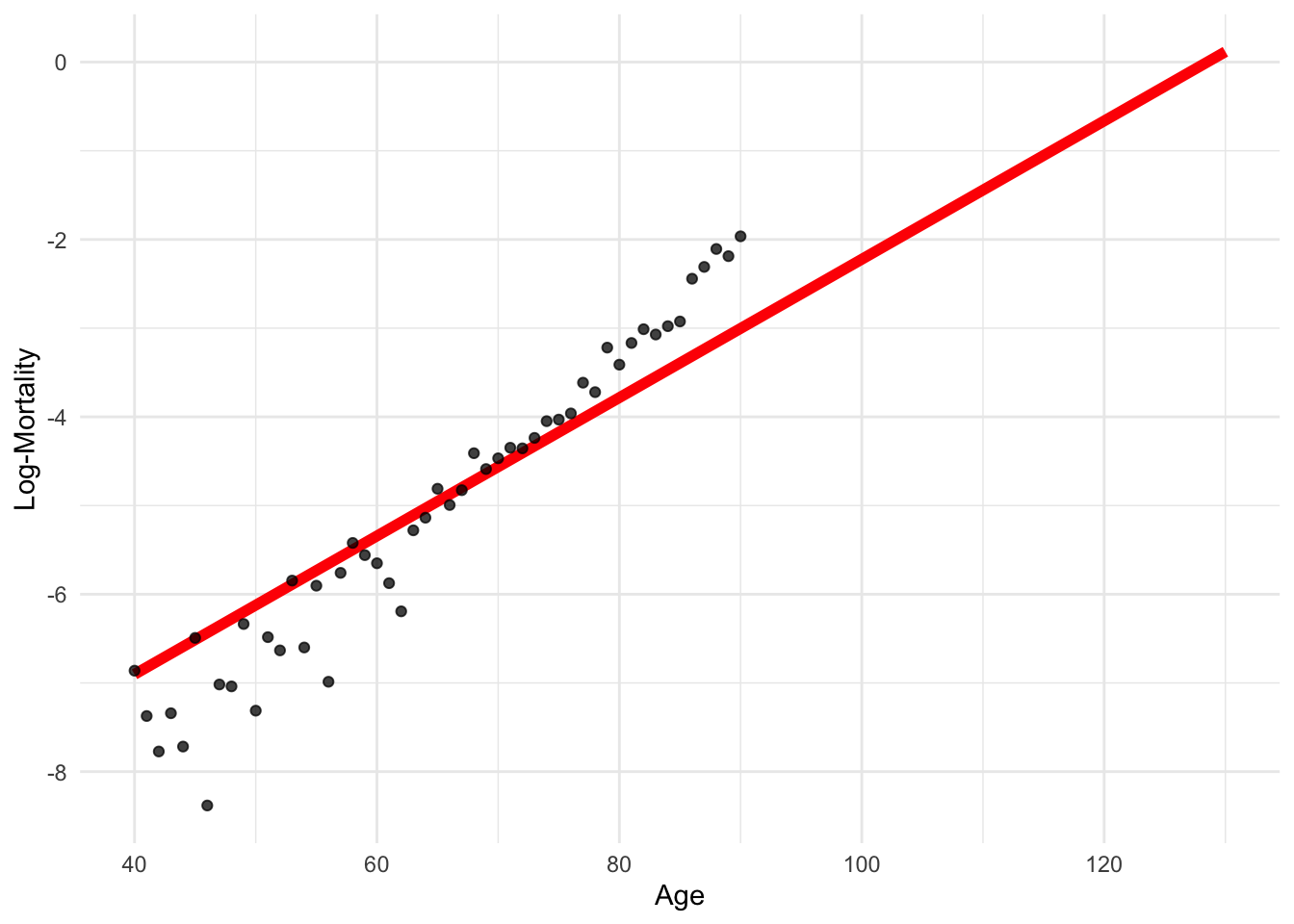
age_grid = pd.DataFrame({"age": np.arange(40, 130)})
predictions = model.predict(age_grid)lux_old = lux_2020[(lux_2020['age'] >= 40) & (lux_2020['age'] <= 130)]
plt.plot(age_grid, model.predict(age_grid), color='red', marker='o')
plt.scatter(lux_old['age'], lux_old['log_mx'], alpha=0.75)
plt.xlabel('Age'); plt.ylabel('Log-Mortality');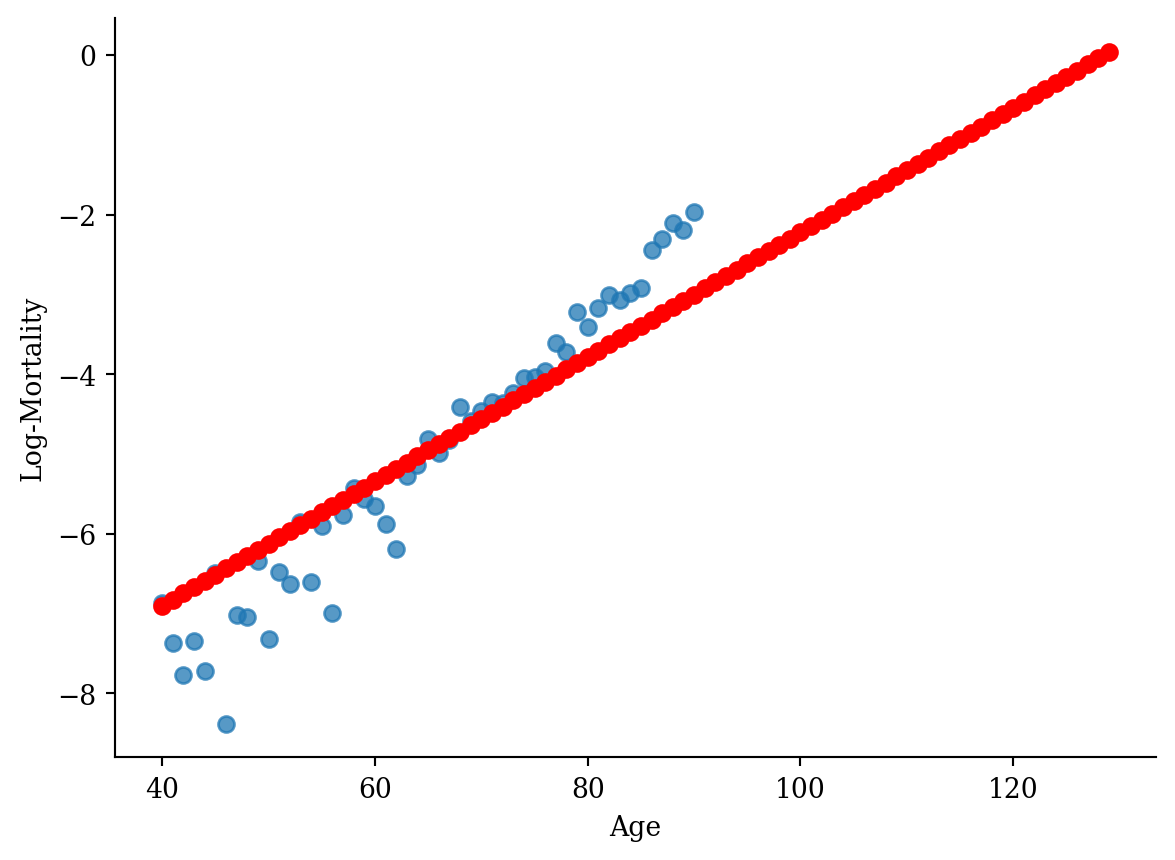
df_mlr = lux[c("age", "year", "log_mx")]
head(df_mlr)# A tibble: 6 × 3
age year log_mx
<int> <int> <dbl>
1 0 1960 -3.74
2 1 1960 -6.38
3 2 1960 -6.37
4 3 1960 -6.68
5 4 1960 -7.08
6 5 1960 -7.04Fitting:
linear_model <- lm(log_mx ~ age + year, data = df_mlr)Predicting:
new_point <- data.frame(year = 2040, age = 20)
predict(linear_model, newdata = new_point) 1
-8.66 coef(linear_model)(Intercept) age year
34.58222358 0.07287039 -0.02191158 # This code chunk is courtesy of ChatGPT.
library(plotly)
model <- lm(log_mx ~ age + year, data = lux)
# Predict values over a grid to create the regression plane
# Create a sequence of ages and years that covers the range of your data
age_range <- seq(min(lux$age), max(lux$age), length.out = 100)
year_range <- seq(min(lux$year), max(lux$year), length.out = 100)
# Create a grid of age and year values
grid <- expand.grid(age = age_range, year = year_range)
# Predict log_mx for the grid
grid$log_mx <- predict(model, newdata = grid)
# Plot
fig <- plot_ly(lux, x = ~age, y = ~year, z = ~log_mx, type = 'scatter3d', mode = 'markers', size = 0.5) %>%
add_trace(data = grid, x = ~age, y = ~year, z = ~log_mx, type = 'mesh3d', opacity = 0.5) %>%
layout(scene = list(xaxis = list(title = 'Age'),
yaxis = list(title = 'Year'),
zaxis = list(title = 'Log-Mortality')))
# Show the plot
fig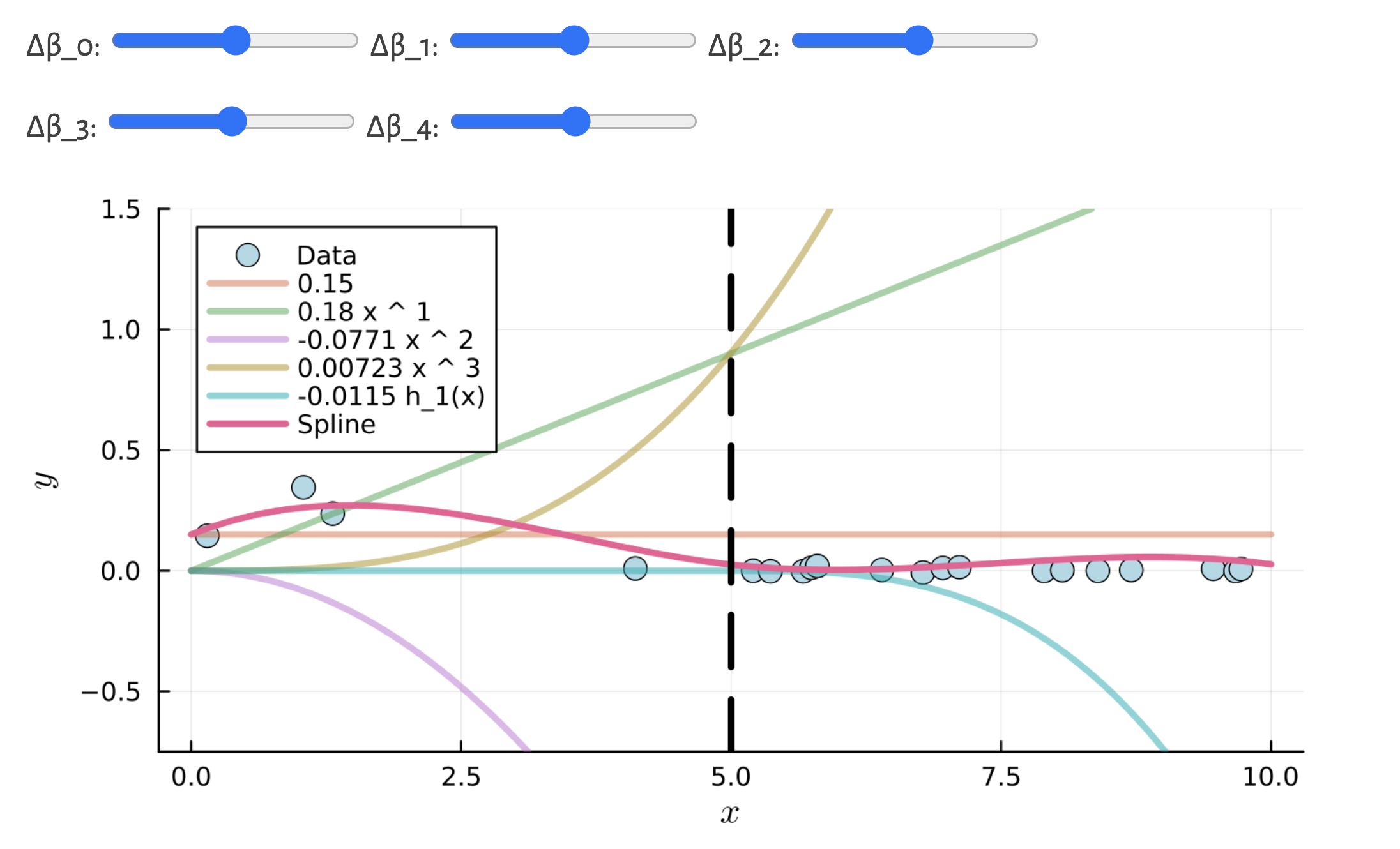
Extend the standard linear model
y_i = \beta_0 + \beta_1 x_i + \varepsilon_i
to
y_i = \beta_0 + \beta_1 x_i + \beta_2 x_i^2 + \cdots + \beta_d x_i^d + \varepsilon_i
df_pr <- data.frame(age = lux_2020$age, age2 = lux_2020$age^2, log_mx = lux_2020$log_mx)
head(df_pr) age age2 log_mx
1 0 0 -5.363176
2 6 36 -8.111728
3 15 225 -6.949619
4 16 256 -8.040959
5 18 324 -7.389022
6 21 441 -8.159519manual_poly_model <- lm(log_mx ~ age + age2,
data = df_pr)
coef(manual_poly_model) (Intercept) age age2
-7.065977594 -0.066603952 0.001421058 impossible_x <- data.frame(age=20, age2=20)
predict(manual_poly_model, newdata = impossible_x) 1
-8.369635 ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
geom_line(data = df_pr, aes(x = age, y = predict(manual_poly_model)), color = "red", linewidth=2) +
geom_point(alpha = 0.75) + labs(x = "Age", y = "Log-Mortality")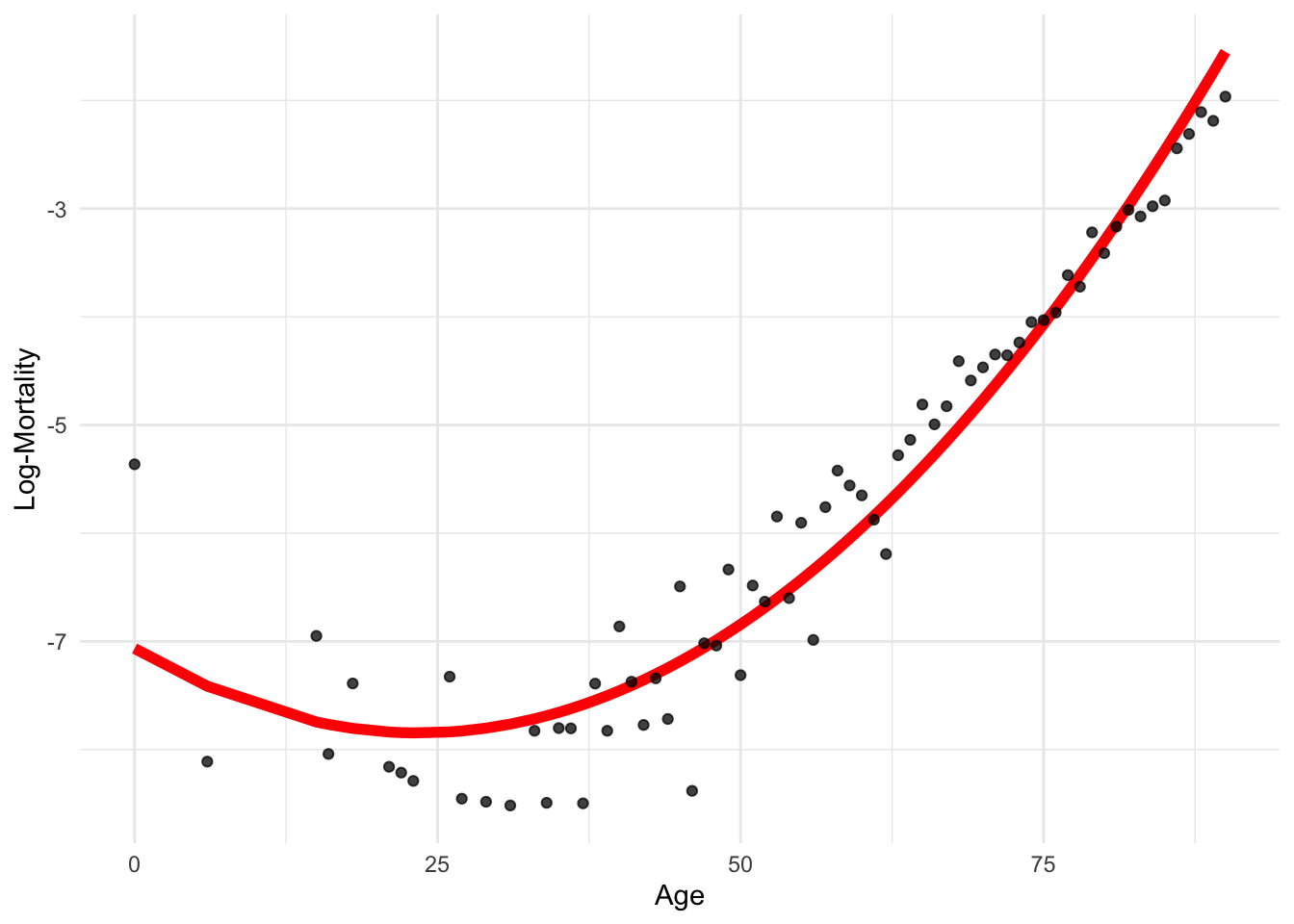
We “tricked” into thinking that age2 is a separate variable!
This is a linear model of a nonlinearly transformed variable.
poly functiondf_pr <- data.frame(age = lux_2020$age, log_mx = lux_2020$log_mx)
head(df_pr) age log_mx
1 0 -5.363176
2 6 -8.111728
3 15 -6.949619
4 16 -8.040959
5 18 -7.389022
6 21 -8.159519poly_model <- lm(log_mx ~ poly(age, 2),
data = df_pr)
coef(poly_model) (Intercept) poly(age, 2)1 poly(age, 2)2
-5.787494 14.534731 6.376355 Now we can’t put in age^2 as a separate variable.
new_input <- data.frame(age = 20)
predict(poly_model, newdata = new_input) 1
-7.829633 ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
geom_line(data = df_pr, aes(x = age, y = predict(poly_model)), color = "red", linewidth=2) +
geom_point(alpha = 0.75) + labs(x = "Age", y = "Log-Mortality")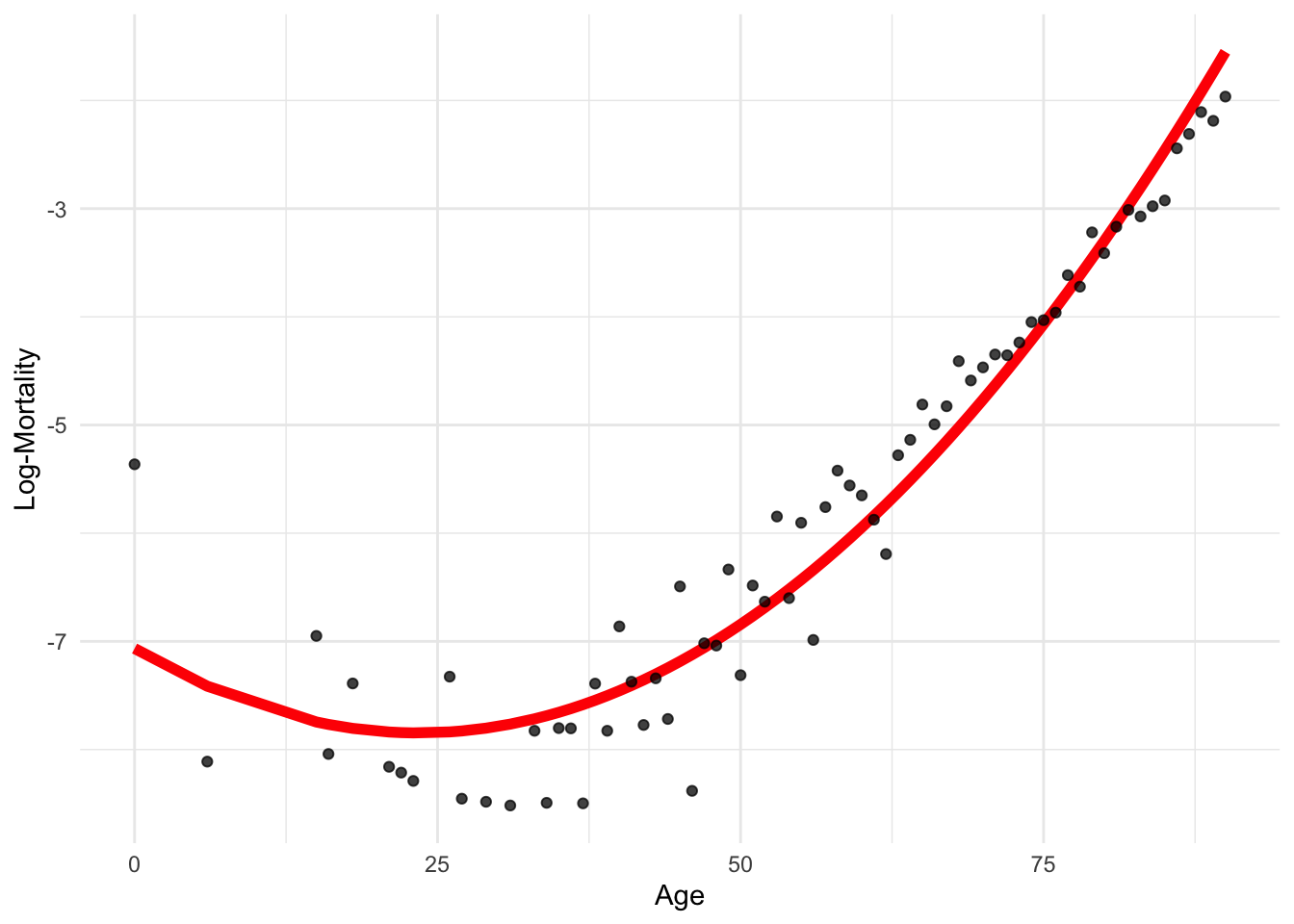
The coefficients are different, but the predictions are the same!
ggplot(lux_2020, aes(x = age, y = log_mx)) + theme_minimal() +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), color = "red", linewidth=2) +
geom_point(alpha = 0.75) + labs(x = "Age", y = "Log-Mortality")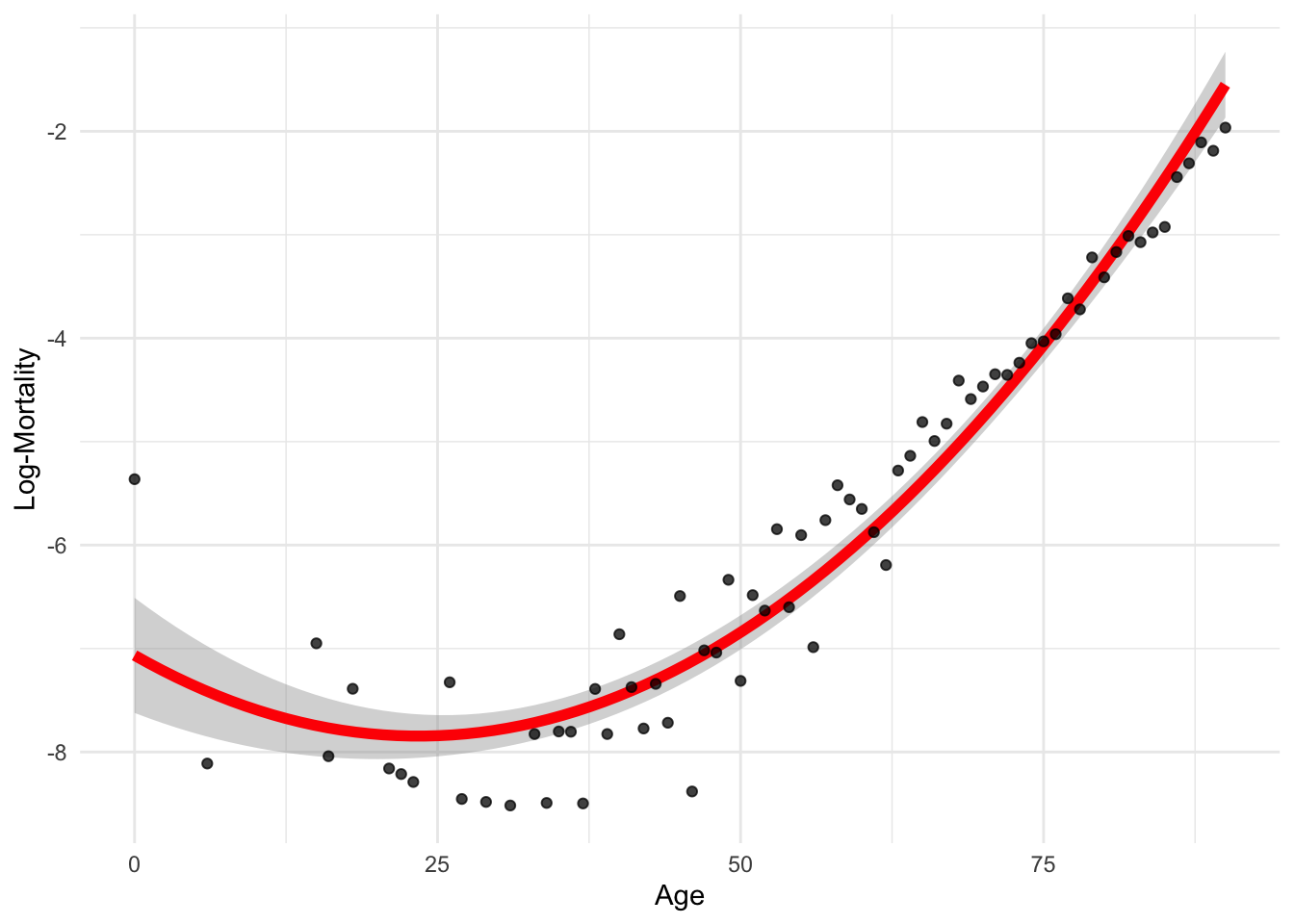
head(lux$age)[1] 0 1 2 3 4 5age_poly <- model.matrix(~ poly(age, 2), data = lux)
head(age_poly) (Intercept) poly(age, 2)1 poly(age, 2)2
1 1 -0.03020513 0.03969719
2 1 -0.02961226 0.03744658
3 1 -0.02901939 0.03524373
4 1 -0.02842652 0.03308866
5 1 -0.02783365 0.03098136
6 1 -0.02724077 0.02892183age_poly <- model.matrix(~ poly(age, 2, raw=TRUE), data = lux)
head(age_poly) (Intercept) poly(age, 2, raw = TRUE)1 poly(age, 2, raw = TRUE)2
1 1 0 0
2 1 1 1
3 1 2 4
4 1 3 9
5 1 4 16
6 1 5 25raw=TRUE)age_poly <- model.matrix(~ poly(age, 2, raw=TRUE), data = lux)df_poly <- data.frame(age = lux$age, age_poly[, -1])
df_poly_long <- df_poly %>%
pivot_longer(cols = -age, names_to = "Polynomial", values_to = "Value")
ggplot(df_poly_long, aes(x = age, y = Value, color = Polynomial)) +
geom_line(linewidth=2) +
theme_minimal() +
labs(title = "Polynomial Terms of Age",
x = "Age",
y = "Polynomial Value",
color = "Term")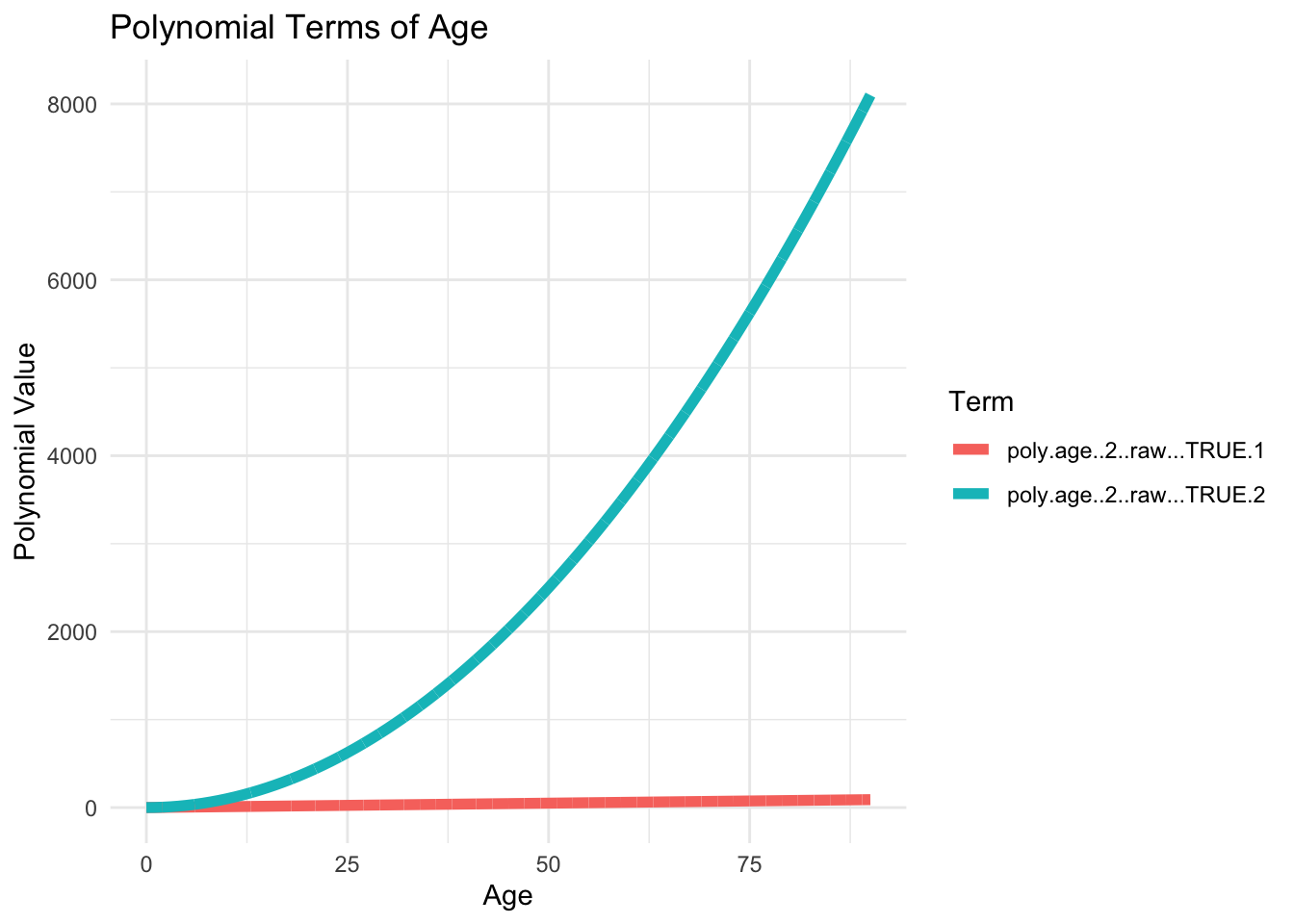
age_poly <- model.matrix(~ poly(age, 4), data = lux)df_poly <- data.frame(age = lux$age, age_poly[, -1])
df_poly_long <- df_poly %>%
pivot_longer(cols = -age, names_to = "Polynomial", values_to = "Value")
ggplot(df_poly_long, aes(x = age, y = Value, color = Polynomial)) +
geom_line(linewidth=2) +
theme_minimal() +
labs(title = "Polynomial Terms of Age",
x = "Age",
y = "Polynomial Value",
color = "Term")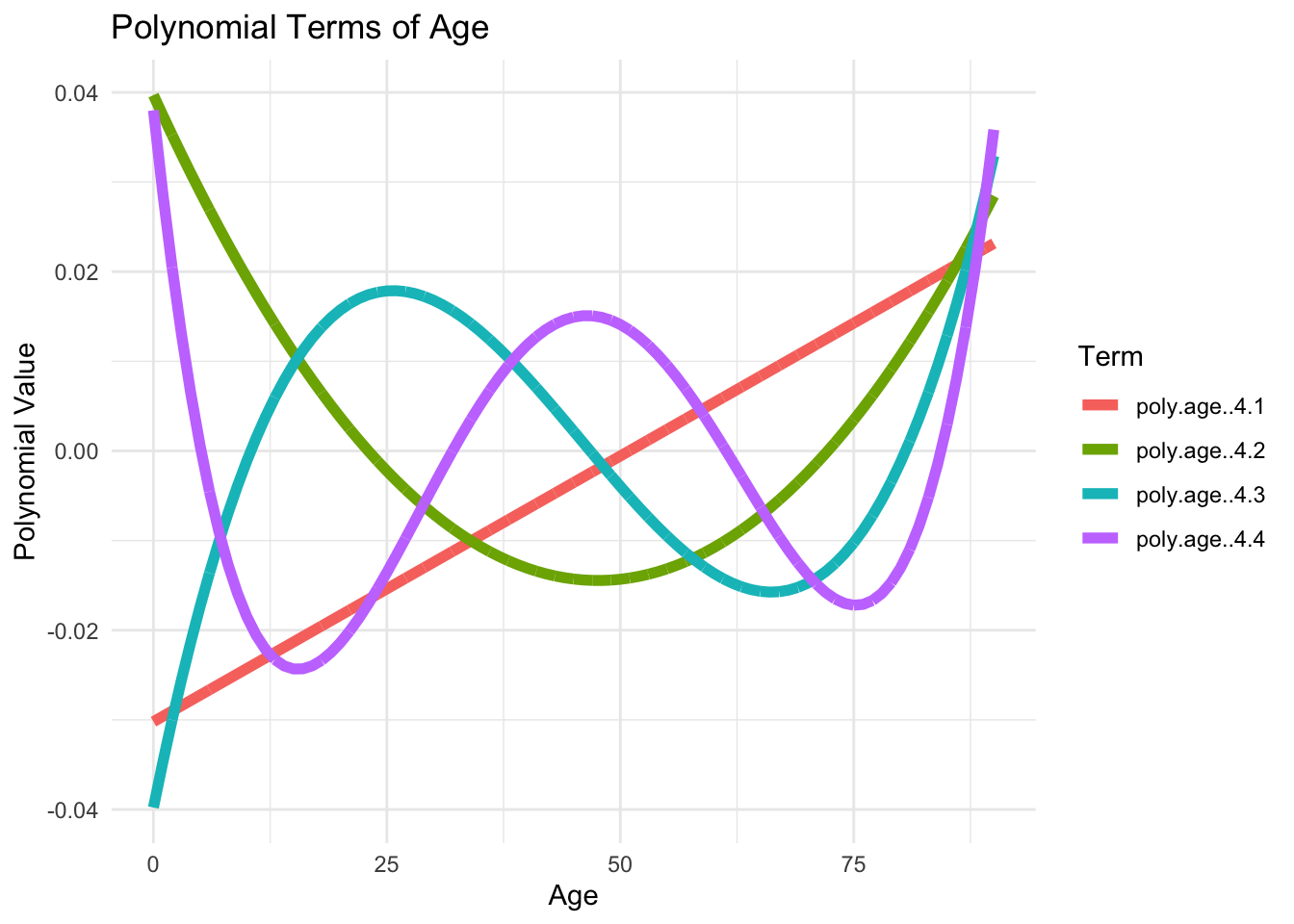
Can you spot an important difference?
summary(manual_poly_model)
Call:
lm(formula = log_mx ~ age + age2, data = df_pr)
Residuals:
Min 1Q Median 3Q Max
-1.25899 -0.33912 -0.03671 0.30503 1.70280
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -7.0659776 0.2787615 -25.348 < 2e-16 ***
age -0.0666040 0.0115898 -5.747 2.43e-07 ***
age2 0.0014211 0.0001108 12.820 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4974 on 67 degrees of freedom
Multiple R-squared: 0.9383, Adjusted R-squared: 0.9364
F-statistic: 509.2 on 2 and 67 DF, p-value: < 2.2e-16summary(poly_model)
Call:
lm(formula = log_mx ~ poly(age, 2), data = df_pr)
Residuals:
Min 1Q Median 3Q Max
-1.25899 -0.33912 -0.03671 0.30503 1.70280
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -5.78749 0.05945 -97.36 <2e-16 ***
poly(age, 2)1 14.53473 0.49736 29.22 <2e-16 ***
poly(age, 2)2 6.37636 0.49736 12.82 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.4974 on 67 degrees of freedom
Multiple R-squared: 0.9383, Adjusted R-squared: 0.9364
F-statistic: 509.2 on 2 and 67 DF, p-value: < 2.2e-16orders <- 1:20
for (i in seq_along(orders)) {
order <- orders[i]
formula <- as.formula(glue("log_mx ~ poly(age, {order})"))
model <- lm(formula, data = lux_2020)
age_grid$log_mx <- predict(model, newdata = age_grid)
plot_regression(lux_2020, age_grid, "age", "log_mx",
glue("Polynomial Regression Order {order}"),
glue("./Animations/NonLinear/Poly{i}.svg"))
}Wage > 250 via logistic regression\mathbb{P}(y_i > 250|x_i) = \frac{\exp(\beta_0 + \beta_1x_i + \beta_2x_i^2 + \beta_3x_i^3 + \beta_4x_i^4)}{1+\exp(\beta_0 + \beta_1x_i + \beta_2x_i^2 + \beta_3x_i^3 + \beta_4x_i^4)}
Pros:
Cons:
Polynomial regression imposes a global structure on the nonlinear function; a simple alternative is to use step functions.
Break up the range of x into k+1 distinct regions (or “bins”), using k cutpoints: c_1 < c_2 < \dots < c_k
Do a least squares fit on y_i = \beta_0 + \beta_1 I(c_1 \leq x_i < c_2) + \beta_2 I(c_2 \leq x_i < c_3) + \dots + \beta_{k-1} I(c_{k-1} \leq x_i < c_k) + \beta_k I(c_k \leq x_i)
Wage dataWage example as before but with step functions.I and cuthead(model.matrix(~ age + I(age >= 3), data = lux)) (Intercept) age I(age >= 3)TRUE
1 1 0 0
2 1 1 0
3 1 2 0
4 1 3 1
5 1 4 1
6 1 5 1head(cut(lux$age, breaks=c(0, 5, 100), right=T))[1] <NA> (0,5] (0,5] (0,5] (0,5] (0,5]
Levels: (0,5] (5,100]head(model.matrix(~ age + cut(age, breaks=c(0, 5, 100), right=T), data = lux)) (Intercept) age cut(age, breaks = c(0, 5, 100), right = T)(5,100]
2 1 1 0
3 1 2 0
4 1 3 0
5 1 4 0
6 1 5 0
7 1 6 1model_step <- lm(log_mx ~ cut(age, breaks=c(0, 5, 100), right=T), data = lux)
coef(model_step) (Intercept)
-7.253560
cut(age, breaks = c(0, 5, 100), right = T)(5,100]
2.011324 breaks be a single numberhead(cut(lux$age, breaks=4, right=T))[1] (-0.09,22.5] (-0.09,22.5] (-0.09,22.5] (-0.09,22.5] (-0.09,22.5]
[6] (-0.09,22.5]
Levels: (-0.09,22.5] (22.5,45] (45,67.5] (67.5,90.1]model_step <- lm(log_mx ~ cut(age, breaks=4, right=T), data = lux)
summary(model_step)
Call:
lm(formula = log_mx ~ cut(age, breaks = 4, right = T), data = lux)
Residuals:
Min 1Q Median 3Q Max
-3.1872 -0.6035 -0.0220 0.5886 3.5764
Coefficients:
Estimate Std. Error t value
(Intercept) -7.24326 0.03111 -232.817
cut(age, breaks = 4, right = T)(22.5,45] 0.14673 0.03896 3.766
cut(age, breaks = 4, right = T)(45,67.5] 1.98093 0.03825 51.783
cut(age, breaks = 4, right = T)(67.5,90.1] 4.37103 0.03797 115.126
Pr(>|t|)
(Intercept) < 2e-16 ***
cut(age, breaks = 4, right = T)(22.5,45] 0.000168 ***
cut(age, breaks = 4, right = T)(45,67.5] < 2e-16 ***
cut(age, breaks = 4, right = T)(67.5,90.1] < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.8284 on 4786 degrees of freedom
Multiple R-squared: 0.8227, Adjusted R-squared: 0.8226
F-statistic: 7404 on 3 and 4786 DF, p-value: < 2.2e-16Fit the model:
y_i = \beta_0 + \beta_1 b_1(x_i) + \beta_2 b_2(x_i) + \dots + \beta_k b_k(x_i)
\boldsymbol{X} = \begin{bmatrix} 1 & x_{11} & x_{12} & \dots & x_{1p} \\ 1 & x_{21} & x_{22} & \dots & x_{2p} \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 1 & x_{n1} & x_{n2} & \dots & x_{np} \end{bmatrix}
\boldsymbol{X} = \begin{bmatrix} 1 & b_1(x_1) & b_2(x_1) & \dots & b_k(x_1) \\ 1 & b_1(x_2) & b_2(x_2) & \dots & b_k(x_2) \\ \vdots & \vdots & \vdots & \ddots & \vdots \\ 1 & b_1(x_n) & b_2(x_n) & \dots & b_k(x_n) \end{bmatrix}
We can combine the previous two ideas (polynomial regression and step functions) to gain flexibility.
For example, you could fit a piecewise cubic polynomial with one “knot”: y_i = \begin{cases} \beta_{0,1} + \beta_{1,1} x_i + \beta_{2,1} x_i^2 + \beta_{3,1} x_i^3 & \text{ if } x_i < c \\ \beta_{0,2} + \beta_{1,2} x_i + \beta_{2,2} x_i^2 + \beta_{3,2} x_i^3 & \text{ if } x_i \geq c \end{cases}
We call c a “knot”: a point of our choosing where the coefficients of the model change.
Question: what are possible issues with this approach?
Wage dataA piecewise polynomial function of degree d is a spline if the function is continuous up to its (d-1)th derivative at each knot.
model <- lm(log_mx ~ bs(age, degree=1, knots=...), data = lux_2020)# Linear regression splines with different number of knots
knots_seq <- 2:11
for (i in seq_along(knots_seq)) {
n_knots <- knots_seq[i]
knots <- quantile(lux_2020$age, probs = seq(0, 1, length.out = n_knots + 1))[-c(1, n_knots + 2)]
formula <- as.formula(glue("log_mx ~ bs(age, degree=1, knots = c({toString(knots)}))"))
model <- lm(formula, data = lux_2020)
age_grid$log_mx <- predict(model, newdata = age_grid)
plot_regression(lux_2020, age_grid, "age", "log_mx",
glue("Cubic Spline with {n_knots} Knots"),
glue("./Animations/NonLinear/SplineCub{i}.svg"),
knots = knots)
}model <- lm(log_mx ~ bs(age, degree=3, knots=...), data = lux_2020)# Cubic regression splines with different number of knots
knots_seq <- 2:11
for (i in seq_along(knots_seq)) {
n_knots <- knots_seq[i]
knots <- quantile(lux_2020$age, probs = seq(0, 1, length.out = n_knots + 1))[-c(1, n_knots + 2)]
formula <- as.formula(glue("log_mx ~ bs(age, degree=3, knots = c({toString(knots)}))"))
model <- lm(formula, data = lux_2020)
age_grid$log_mx <- predict(model, newdata = age_grid)
plot_regression(lux_2020, age_grid, "age", "log_mx",
glue("Cubic Spline with {n_knots} Knots"),
glue("./Animations/NonLinear/SplineCub{i}.svg"),
knots = knots)
}Wage dataThe cubic is preferred as it is the smallest order where the knots are not visible without close inspection.
We can extend the idea of a cubic spline to a natural cubic spline. It is a spline where outside the boundary knots (extrapolation) the function is linear.
Wage subsetWage data\text{minimise:} \quad \frac{1}{n} \sum_{i=1}^n (y_i - g(x_i))^2 + \lambda \int g''(x)^2 \, \mathrm{d}x.
For mortality smoothing, US uses smoothing splines, UK uses regression splines.
model <- smooth.spline(lux_2020$age, lux_2020$log_mx, df = df)# Smoothing splines with different df (degrees of freedom)
dfs <- seq(2, 20, length.out = 10)
for (i in seq_along(dfs)) {
df <- dfs[i]
model <- smooth.spline(lux_2020$age, lux_2020$log_mx, df = df)
predictions <- predict(model, age_grid$age)
age_grid$log_mx <- predictions$y
plot_regression(lux_2020, age_grid, "age", "log_mx",
glue("Smoothing Spline with df {df}"),
glue("./Animations/NonLinear/SmoothSpline{i}.svg"))
}cv.spline <- smooth.spline(lux_2020$age, lux_2020$log_mx, cv=TRUE)
cv.spline$df[1] 5.08017plot(lux_2020$age, lux_2020$log_mx, xlab="age", ylab="log_mx", pch=19)
lines(cv.spline, col="brown3", lwd=3)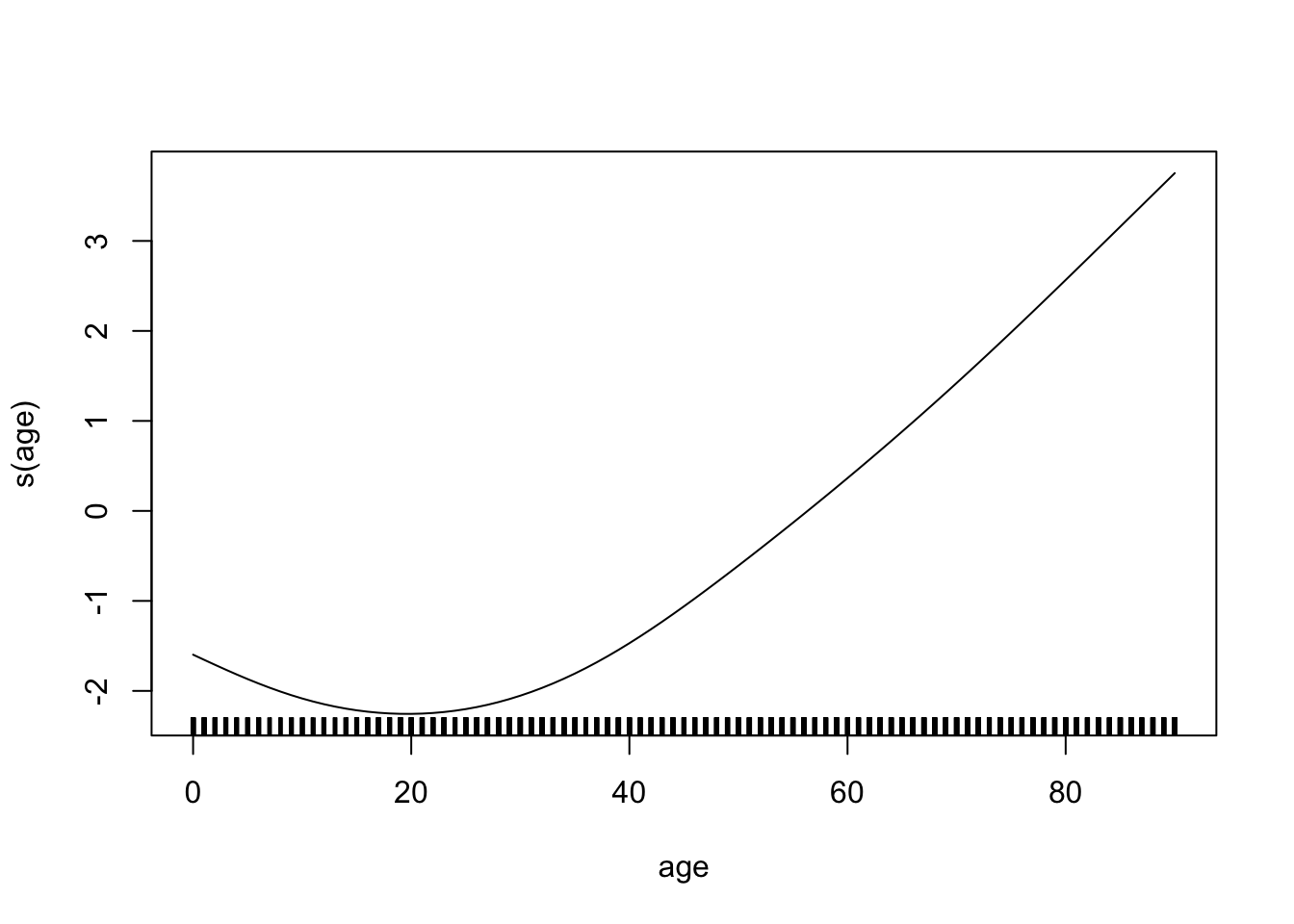
Wage dataIdea: predict the response y_0 associated with predictor x_0 via a linear fit, but using only the data points (x_i,y_i) that are “close” to x_0.
Algorithm:
Wage datay_i = \beta_0 + f_1(x_{i,1}) + f_2(x_{i,2}) + ... + f_p(x_{i,p}) + \varepsilon_i
Wage datawage.education is qualitative. The others are fit with natural cubic splines.Wage datawage.education is qualitative. The others are fit with smoothing splines.mgcv package)library(mgcv)
model_gam <- gam(log_mx ~ s(age) + s(year), data=lux)plot(model_gam, select=1)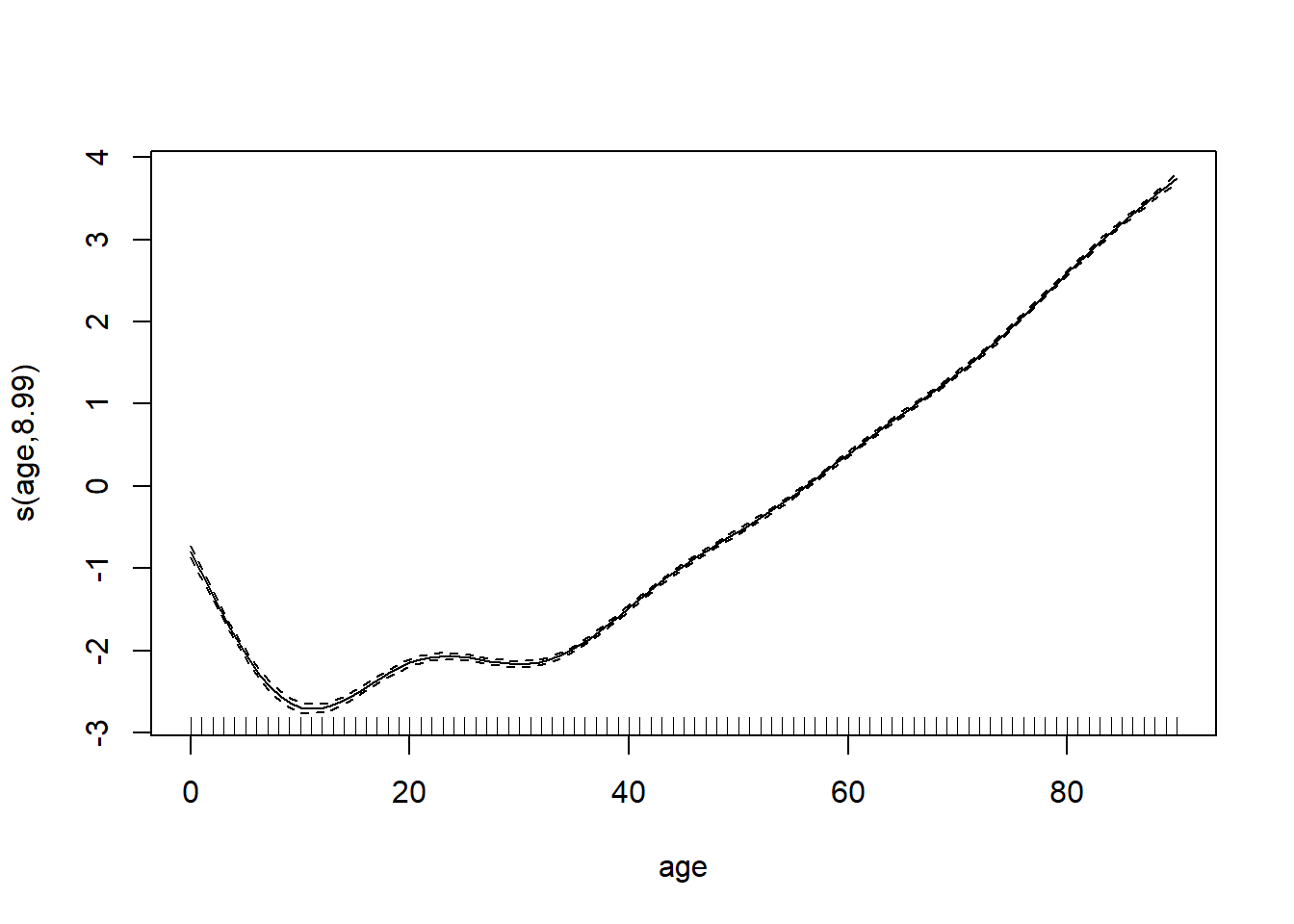
plot(model_gam, select=2)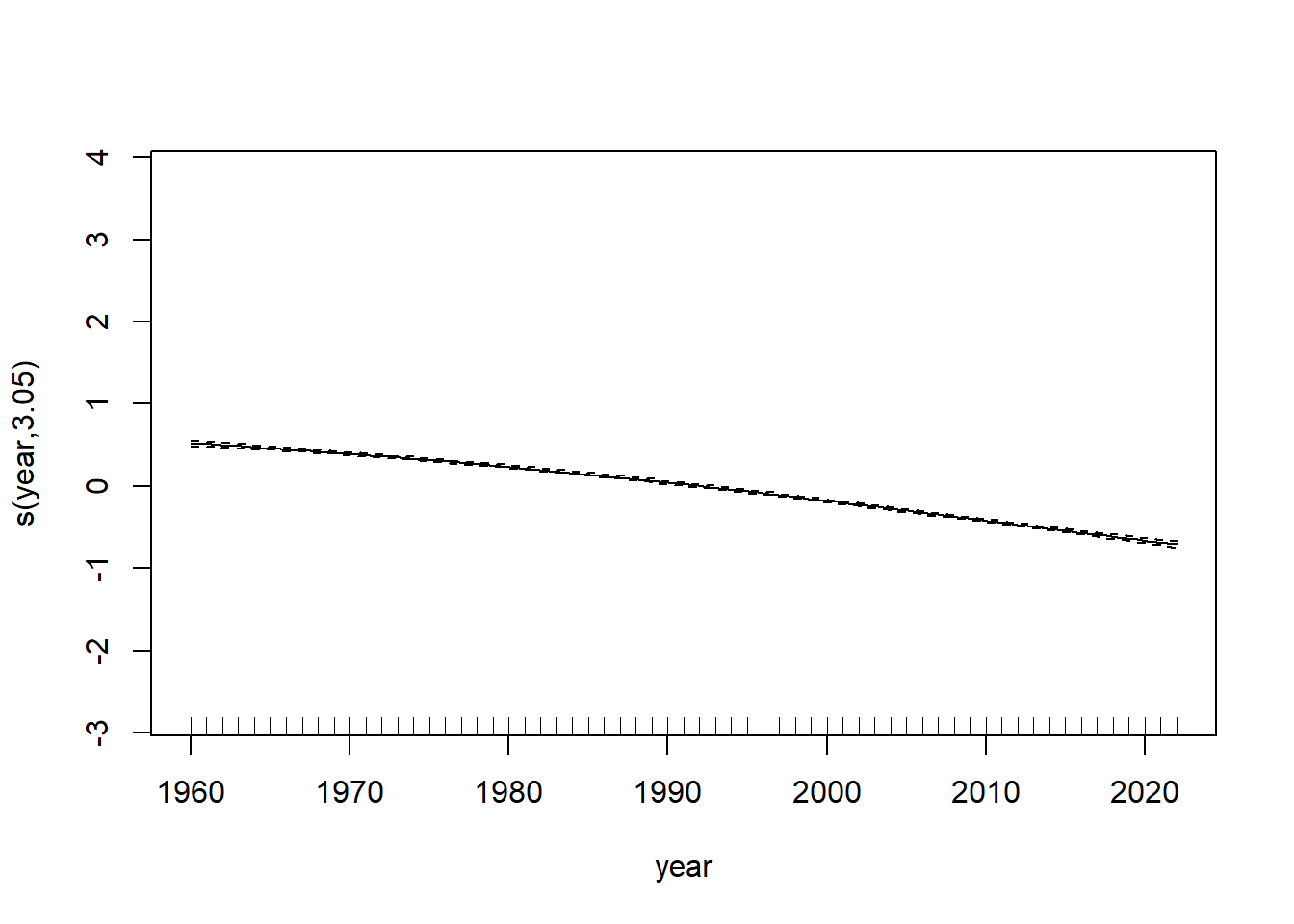
gam package)library(gam)
lux_factor <- lux %>% mutate(year = factor(year))
model_gam <- gam(log_mx ~ s(age) + year, data=lux_factor)
plot(model_gam)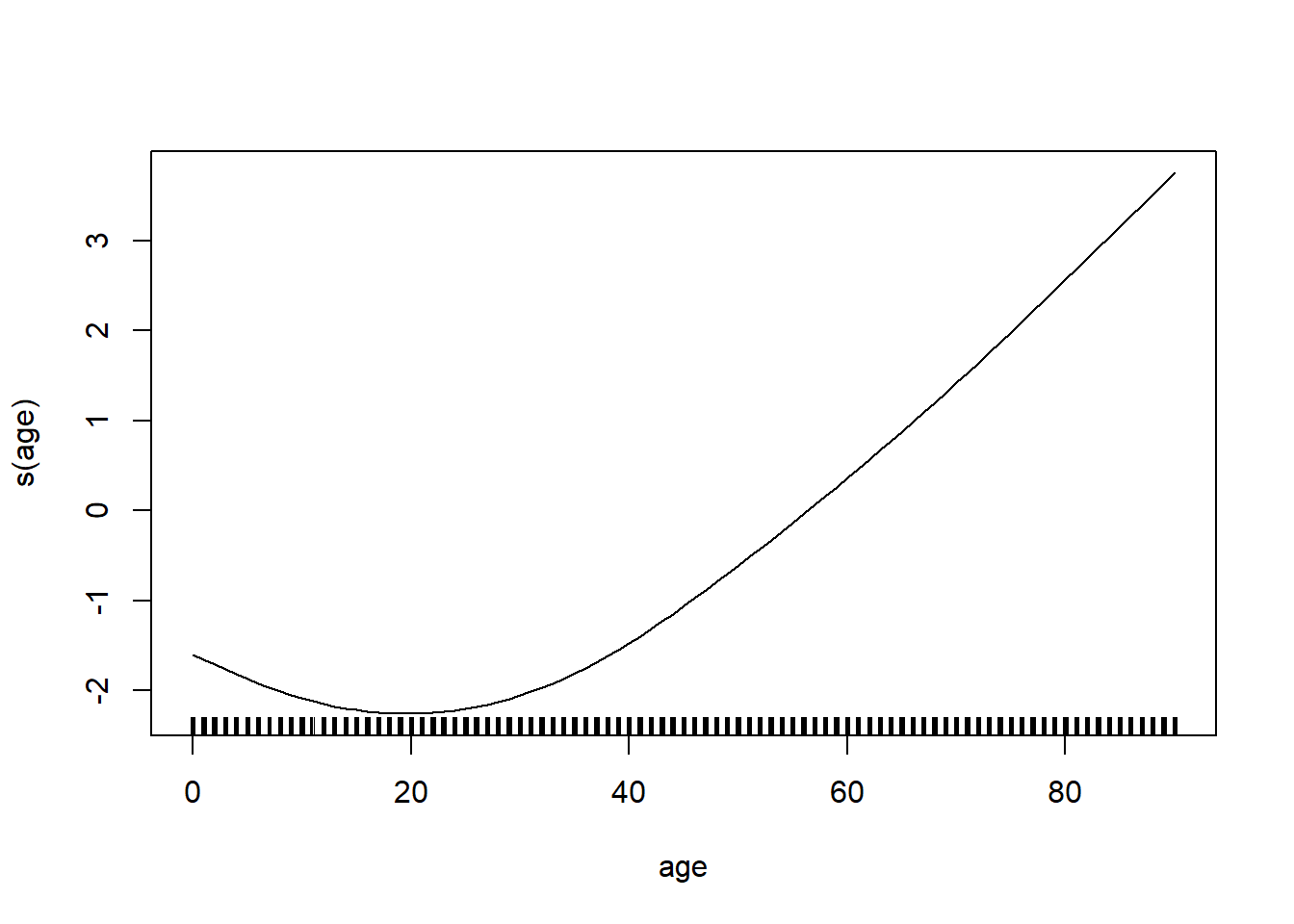
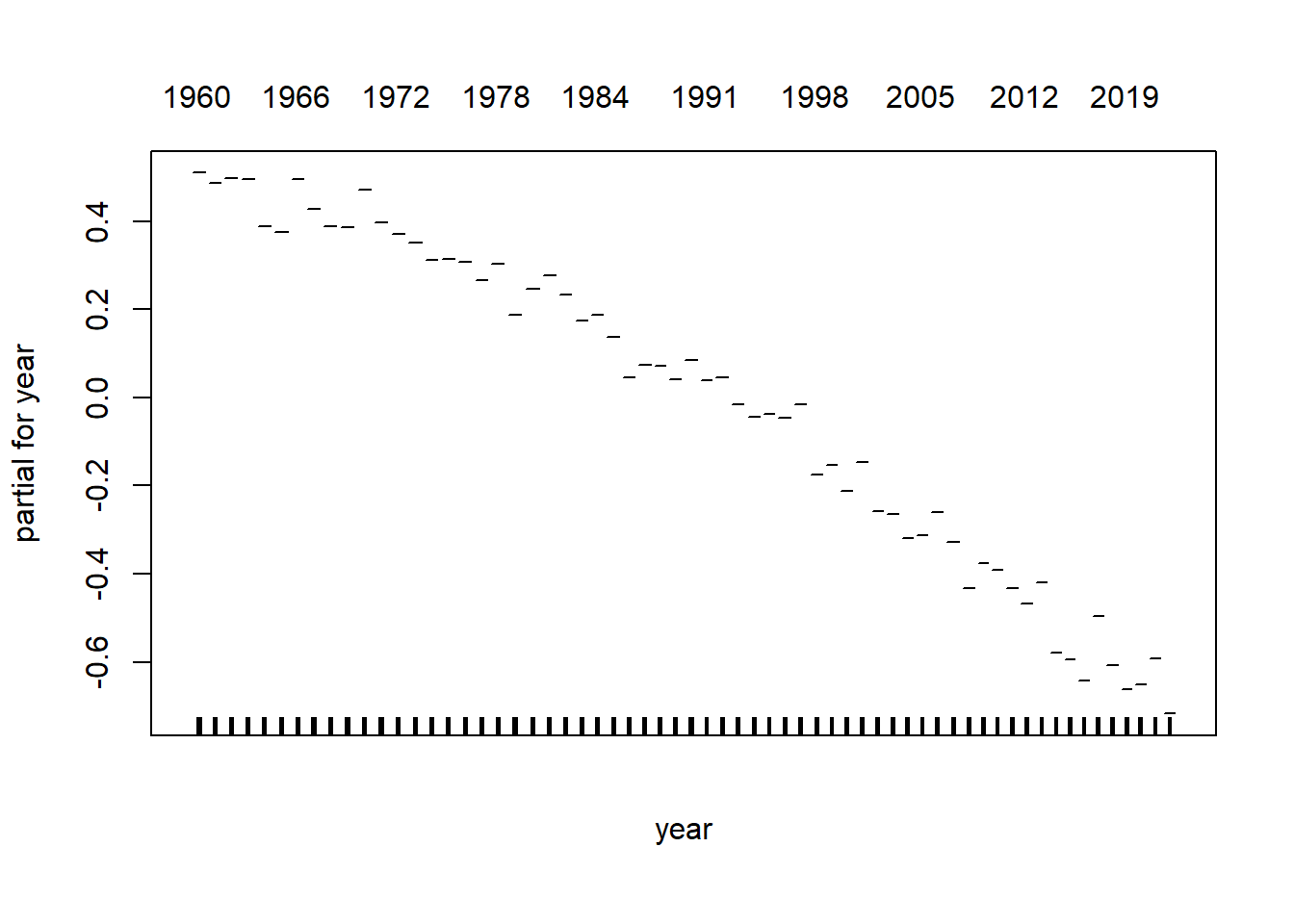
It won’t help with your assessment, it’s just very entertaining/interesting.
print(sessionInfo(), locale=FALSE, tzone=FALSE)R version 4.5.0 (2025-04-11 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
LAPACK version 3.12.1
attached base packages:
[1] splines stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] gam_1.22-5 foreach_1.5.2 glue_1.8.0 plotly_4.10.4
[5] mgcv_1.9-1 nlme_3.1-168 lubridate_1.9.4 forcats_1.0.0
[9] stringr_1.5.1 dplyr_1.1.4 purrr_1.0.4 readr_2.1.5
[13] tidyr_1.3.1 tibble_3.2.1 ggplot2_3.5.2 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] generics_0.1.4 stringi_1.8.7 lattice_0.22-6 hms_1.1.3
[5] digest_0.6.37 magrittr_2.0.3 evaluate_1.0.3 grid_4.5.0
[9] timechange_0.3.0 RColorBrewer_1.1-3 iterators_1.0.14 fastmap_1.2.0
[13] rprojroot_2.0.4 jsonlite_2.0.0 Matrix_1.7-3 httr_1.4.7
[17] crosstalk_1.2.1 viridisLite_0.4.2 scales_1.4.0 codetools_0.2-20
[21] lazyeval_0.2.2 cli_3.6.5 rlang_1.1.6 withr_3.0.2
[25] yaml_2.3.10 tools_4.5.0 tzdb_0.5.0 here_1.0.1
[29] reticulate_1.42.0 vctrs_0.6.5 R6_2.6.1 png_0.1-8
[33] lifecycle_1.0.4 htmlwidgets_1.6.4 pkgconfig_2.0.3 pillar_1.10.2
[37] gtable_0.3.6 data.table_1.17.2 Rcpp_1.0.14 xfun_0.52
[41] tidyselect_1.2.1 knitr_1.50 farver_2.1.2 htmltools_0.5.8.1
[45] rmarkdown_2.29 labeling_0.4.3 compiler_4.5.0 from watermark import watermark
print(watermark(python=True, packages="matplotlib,numpy,pandas,seaborn,scipy"))Python implementation: CPython
Python version : 3.11.12
IPython version : 9.2.0
matplotlib: 3.10.3
numpy : 2.2.6
pandas : 2.2.3
seaborn : 0.13.2
scipy : 1.15.3